MS facilities for quantitative analysis
Mark Bennett is the lab manager of the MS facilities for quantitative analysis within the Department of Life Sciences.
Our analytical equipment includes both LC-MS/MS and GC-MS platforms.
The facility handles a very wide range of projects, covering both proteins and small molecules. Recently we have been focussing on methods for targeted quantitative measurements of tryptic peptides (“Mass Westerns”) as an alternative to using antibodies for quantifying proteins. Further examples of the types of analysis available are listed below.
- Quantitative proteomics including targeted analysis and quantitative studies of post translational modifications.
- Analysis of plant hormones from Arabidopsis and other species.
- Sugar analysis from plants and aphids.
- Analysis of glucosinolates and their degradation products.
- Fatty acid profiling of microbes and plants.
- DNA/RNA oxidation in microbes.
- Analysis of cyclic dinucleotides in bacteria.
- Proteomic analysis of Arabidopsis and other plants.
details
LCMS
The core LC-MS systems comprises of two highly flexible ABSCIEX QTrap LCMS instruments. The QTrap MS is a hybrid Triple Quadrupole/Ion Trap mass spectrometer that can be used in several modes depending on the application.

Our most sensitive instrument, the latest 6500 model, was recently purchased with college SIF funding; the MS is coupled to an Eksigent microflow LC system to provide a very sensitive but robust platform for high throughput analysis of peptides and small molecules.
An older 2000QTrap is available for the analysis of molecules where the greatest sensitivity is not required, this can coupled to Agilent binary or capillary LC systems.
Several other standalone LC systems are available these are equipped with a range of detectors (UV, RI,DAD).
How it works
The MS has three quadrupoles: Q1 and Q3 are used as mass filters; Q2 is a collision cell where intact molecules and fragments from Q1 are broken into fragments of smaller masses.
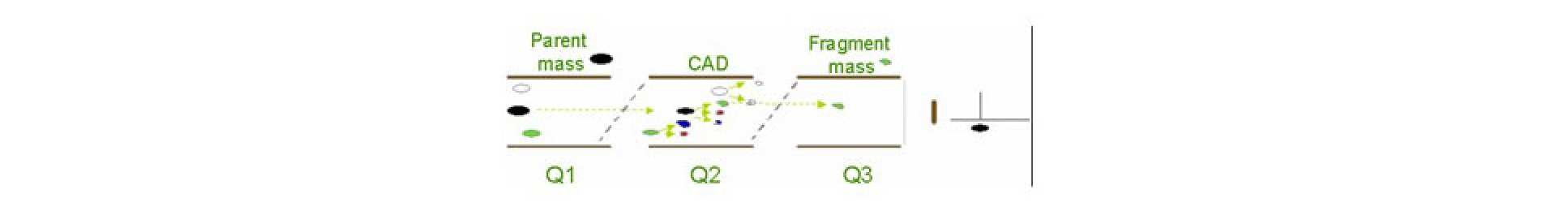
Triple Quad modes
MRM (multiple reaction monitoring) - Q1 and Q3 fixed: Q3 set to detect only one particular mass that is a known fragment (daughter) from the intact molecule (parent) of known mass filtered through Q1.
This is the standard quantitative mode used for targeted metabolite and protein analysis.
Advantages: high sensitivity, high reproducibility, low noise, simultaneous measurement of up to (1000) transitions.
Disadvantages: Only works for known molecules, requires standard compound for full optimisation
Precursor ion scanning - Q3 fixed Q1 scans
Looks for all possible molecules containing a common sub-structure of known mass, an example would be for looking for phosphopeptides in a tryptic digest of a phosphorulated protein by looking for ions showing loss of phosphate.
Neutral loss - Q1 and Q3 scan with a fixed mass difference
Looks for loss of a common fragment occurring in a range of parent compounds.
Ion Trap modes
Enhanced MS (EMS) - Q1 is set to “open” so no mass filtering. Full scan spectra are generated from scans out of Q3 .This is usually used to give an overview of all of the ions in sample.
Enhanced production (EPI) - Q1 is set to a fixed mass (m/z value), usually based on a known molecule. As with EMS, spectra are generated from scans out of Q3. Advantage is to generate full scan spectra from a single parent ion e.g. for peptide sequencing. Sensitivity can approach that achieved via MRM, but EPI provides much more spectral information.
Enhanced resolution (ER) - More accurate m/z determination (but still not as accurate as some other LCMS systems such as QTOF).
Mixed modes (quadrupole/ion trap) are possible. This can be data dependent, for example, a signal from an MRM scan could trigger an EPI scan to confirm a compound identity. Another example would be the selection of only doubly charged molecules to trigger an EPI, as in the analysis of tryptic peptide fragments.
For further information or to discuss access, please contact mhbennett@imperial.ac.uk
GCMS
For GC-MS we operate two Agilent 6890/5973 systems.
The two 6890/5973 Agilent GC/MS systems are single quadrupole instruments. Both systems are set up for split/splitless injection with capillary GC, and operate in electron impact (EI) mode. One of the GCs is additionally equipped with a FID detector, the other has an OPTIC-2 thermal desorption(TD) inlet. TD is largely used for analysis of highly volatile compounds, collected as “headspace” samples adsorbed onto a chemical trap. Biological volatiles include attractants, deterrents and signalling compounds
For further information or to discuss access, please contact mhbennett@imperial.ac.uk
Publications
2017
Sood, D., Johnson, N., Jain, P., Siskos, A. P., Bennett, M., Gilham, C., . . . Fletcher, O. (2017). CYP3A7*1C allele is associated with reduced levels of 2-hydroxylation pathway oestrogen metabolites.. Br J Cancer, 116(3), 382-388. doi:10.1038/bjc.2016.432
Siskos, A. P., Jain, P., Romisch-Margl, W., Bennet, M., Achaintre, D., Asad, Y., . . . Keun, H. C. (2017). Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma. ANALYTICAL CHEMISTRY, 89(1), 656-665. doi:10.1021/acs.analchem.6b02930
2016
Lopez-Cobollo, R. M., Filippis, I., Bennett, M. H., & Turnbull, C. G. N. (2016). Comparative proteomics of cucurbit phloem indicates both unique and shared sets of proteins. PLANT JOURNAL, 88(4), 633-647. doi:10.1111/tpj.13288
Goncalves, S. F., Davies, S. K., Bennett, M., Raab, A., Feldmann, J., Kille, P., . . . Bundy, J. G. (2016). Sub-lethal cadmium exposure increases phytochelatin concentrations in the aquatic snail Lymnaea stagnalis. SCIENCE OF THE TOTAL ENVIRONMENT, 568, 1054-1058. doi:10.1016/j.scitotenv.2016.06.149
Luang-In, V., Albaser, A. A., Nueno-Palop, C., Bennett, M. H., Narbad, A., & Rossiter, J. T. (2016). Glucosinolate and Desulfo-glucosinolate Metabolism by a Selection of Human Gut Bacteria. CURRENT MICROBIOLOGY, 73(3), 442-451. doi:10.1007/s00284-016-1079-8
Albaser, A., Kazana, E., Bennett, M. H., Cebeci, F., Luang-In, V., Spanu, P. D., & Rossiter, J. T. (2016). Discovery of a Bacterial Glycoside Hydrolase Family 3 (GH3) beta-Glucosidase with Myrosinase Activity from a Citrobacter Strain Isolated from Soil. JOURNAL OF AGRICULTURAL AND FOOD CHEMISTRY, 64(7), 1520-1527. doi:10.1021/acs.jafc.5b05381
Stewart, S. A., Hodge, S., Bennett, M., Mansfield, J. W., & Powell, G. (2016). Aphid induction of phytohormones in Medicago truncatula is dependent upon time post-infestation, aphid density and the genotypes of both plant and insect. ARTHROPOD-PLANT INTERACTIONS, 10(1), 41-53. doi:10.1007/s11829-015-9406-8
2015
Luang-In, V., Narbad, A., Cebeci, F., Bennett, M., & Rossiter, J. T. (2015). Identification of Proteins Possibly Involved in Glucosinolate Metabolism in L-agilis R16 and E-coli VL8. PROTEIN JOURNAL, 34(2), 135-146. doi:10.1007/s10930-015-9607-0
2014
Lougheed, K. E. A., Bennett, M. H., & Williams, H. D. (2014). An in vivo crosslinking system for identifying mycobacterial protein-protein interactions. JOURNAL OF MICROBIOLOGICAL METHODS, 105, 67-71. doi:10.1016/j.mimet.2014.07.012
Schumacher, J., Waite, C. J., Bennett, M. H., Perez, M. F., Shethi, K., & Buck, M. (2014). Differential secretome analysis of Pseudomonas syringae pv tomato using gel-free MS proteomics. FRONTIERS IN PLANT SCIENCE, 5, 11 pages. doi:10.3389/fpls.2014.00242
Young, N. F., Ferguson, B. J., Antoniadi, I., Bennett, M. H., Beveridge, C. A., & Turnbull, C. G. N. (2014). Conditional Auxin Response and Differential Cytokinin Profiles in Shoot Branching Mutants. PLANT PHYSIOLOGY, 165(4), 1723-1736. doi:10.1104/pp.114.239996
Engl, C., Waite, C. J., McKenna, J. F., Bennett, M. H., Hamann, T., & Buck, M. (2014). Chp8, a Diguanylate Cyclase from Pseudomonas syringae pv. Tomato DC3000, Suppresses the Pathogen-Associated Molecular Pattern Flagellin, Increases Extracellular Polysaccharides, and Promotes Plant Immune Evasion. MBIO, 5(3), 11 pages. doi:10.1128/mBio.01168-14
2013
Liebeke, M., Garcia-Perez, I., Anderson, C. J., Lawlor, A. J., Bennett, M. H., Morris, C. A., . . . Bundy, J. G. (2013). Earthworms Produce phytochelatins in Response to Arsenic. PLOS ONE, 8(11), 13 pages. doi:10.1371/journal.pone.0081271
Schumacher, J., Behrends, V., Pan, Z., Brown, D. R., Heydenreich, F., Lewis, M. R., . . . Buck, M. (2013). Nitrogen and Carbon Status Are Integrated at the Transcriptional Level by the Nitrogen Regulator NtrC In Vivo. MBIO, 4(6), 9 pages. doi:10.1128/mBio.00881-13
Luang-In, V., Narbad, A., Nueno-Palop, C., Mithen, R., Bennett, M., & Rossiter, J. T. (2014). The metabolism of methylsulfinylalkyl- and methylthioalkyl- glucosinolates by a selection of human gut bacteria. MOLECULAR NUTRITION & FOOD RESEARCH, 58(4), 875-883. doi:10.1002/mnfr.201300377
Fones, H. N., Eyles, C. J., Bennett, M. H., Smith, J. A. C., & Preston, G. M. (2013). Uncoupling of reactive oxygen species accumulation and defence signalling in the metal hyperaccumulator plant Noccaea caerulescens. NEW PHYTOLOGIST, 199(4), 916-924. doi:10.1111/nph.12354
Westwood, J. H., McCann, L., Naish, M., Dixon, H., Murphy, A. M., Stancombe, M. A., . . . Carr, J. P. (2013). A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. MOLECULAR PLANT PATHOLOGY, 14(2), 158-170. doi:10.1111/j.1364-3703.2012.00840.x
Williams, K. J., Bennett, M. H., Barton, G. R., Jenkins, V. A., & Robertson, B. D. (2013). Adenylylation of mycobacterial Glnk (PII) protein is induced by nitrogen limitation. TUBERCULOSIS, 93(2), 198-206. doi:10.1016/j.tube.2012.12.003
Hodge, S., Ward, J. L., Beale, M. H., Bennett, M., Mansfield, J. W., & Powell, G. (2013). Aphid-induced accumulation of trehalose in Arabidopsis thaliana is systemic and dependent upon aphid density. PLANTA, 237(4), 1057-1064. doi:10.1007/s00425-012-1826-4


