New chemical proteomics methods paper in Nature Protocols
Comprehensive Tate group protocols for chemical proteomic analysis have been published in Nature Protocols
The Tate group have published a new paper in Nature Protocols from research led by current group member Wouter Kallemeijn, and group alumni Tom Lanyon-Hogg, Kwang Panyain, Andrea Goya Grocin, Paulina Ciepla and Julia Morales-Sanfrutos.
Protein lipidation is among the most widespread post-translational modifications (PTMs) found in nature, regulating protein function, structure, and subcellular localisation. Lipid transferases and their substrate proteins are also attracting increasing interest as drug targets due to their dysregulation in many disease states. However, the inherent hydrophobicity and potential dynamic nature of lipid modifications makes them notoriously challenging to detect by many analytical methods.
Chemical proteomics provides a powerful approach to identify and quantify these diverse protein modifications by combining bespoke chemical tools for lipidated protein enrichment with quantitative mass spectrometry-based proteomics.
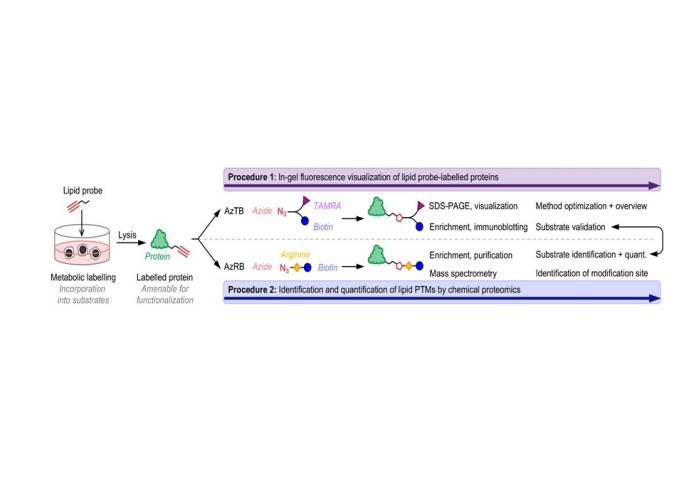
In this Nature Protocols paper, we report a robust proteome-wide approach for the exploration of the five major classes of protein lipidation in living cells – N-myristoylation, N- and S-acylation, O-cholesterylation, S-farnesylation and S-geranylgeranylation – through the use of specific chemical probes for each lipid PTM. In-cell labelling of lipidated proteins is achieved by metabolic incorporation of a lipid probe that mimics the specific natural lipid, carrying an alkyne as bioorthogonal labelling tag. Chemically tagged proteins can be coupled to multifunctional ‘capture reagents’ using click chemistry, allowing in-gel fluorescence visualisation or enrichment via affinity handles for quantitative chemical proteomics based on label-free quantification (LFQ) or tandem mass-tag (TMT) approaches.
Our protocol provides detailed workflows for method optimization, sample preparation for chemical proteomics and data processing, enabling a technician, graduate student, or postdoc to complete all steps from optimizing metabolic labelling to data processing within three weeks. Ultimately, this protocol enables the sensitive and quantitative analysis of lipidated proteins at a proteome-wide scale at native expression levels, which is critical to understanding the role of lipid PTMs in health and disease.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Edward Bartlett
Department of Chemistry