Our KLK6 activity-based probes now published in JACS!
New research from the Tate group developing activity-based probes has recently been published.
Our work on the development of selective activity-based probes for the secreted protease Kallikrein-related peptidase 6 is now out in the Journal of the American Chemical Society.
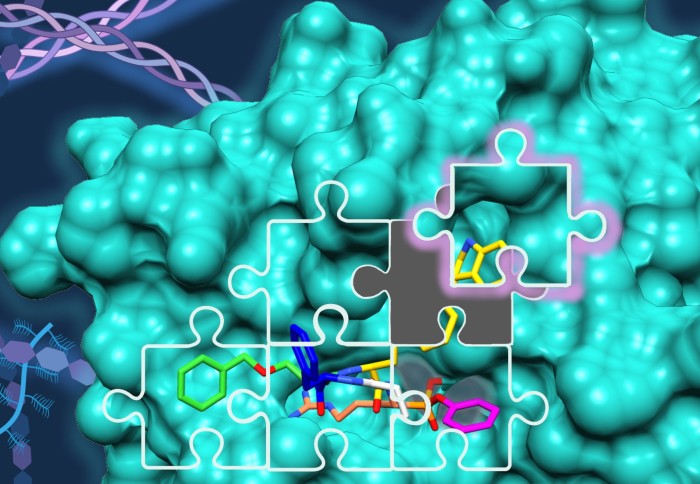
Kallikrein-related peptidases (KLKs) are a family of secreted serine proteases that play an important function in the remodelling of the extracellular matrix in physiology and disease. Their potential as biomarkers is best exemplified by the Prostate Specific Antigen (PSA or KLK3), which is routinely used in the diagnosis of prostate cancer. KLKs activity is tightly controlled by protease-regulated activation and inactivation, a cascade known as the “KLK activome”. Hence to understand these proteases and their role in physiology and disease it is important to assess not only their expression levels but also their activity, an aspect which requires specific chemical tools named activity-based probes (ABP).
Building up on our work on KLK2, KLK3 and KLK14 published last year on JACS, we have now developed the first selective and highly potent ABPs for KLK6 for the study of this KLK’s role in pancreatic ductal adenocarcinoma (PDAC). These probes were optimised by modular design using our in-house positional scanning substrate library for trypsin-like serine proteases, and were found to have exquisite selectivity for KLK6 over KLK8, another highly homologous protease highly expressed in PDAC. The report includes structural biology studies of the ABP binding mode, led by Pravin Kumar Ankush Jagtapat at the European Molecular Biology Laboratory (EMBL) in Heidelberg and Dr Svend Kjær at the Francis Crick Institute.
KLK6 ABPs were validated by proteomics and cellular assays, revealing a clear role for KLK6 activity in the increased migration of PDAC. Furthermore, we were able to show that despite being found in different types of cancer cell models of PDAC, the activity of KLK6 does not usually correlate to its expression levels, highlighting the need for ABPs to really understand this target. We further explored the use of our ABPs to profile KLK6’s substrates in PDAC by employing the Hunter N-terminomic approach, which led to the identification of previously unknown substrates that could shed light on the mechanisms by which KLK6 promotes cancer migration.
The study is the result of a multidisciplinary collaboration with Dr Aubry Miller at the German Cancer Research Centre and was funded by Cancer Research UK (C24523/A25192, C29637/A20183, and C29637/A21451), the EPSRC Centre for Doctoral Training in Physical Sciences Innovation in Chemical Biology for Bioindustry and Healthcare (EP/LO15498/1), the European Union Horizon 2020 Program (MSCA-IF fellowship 890900), and Worldwide Cancer Research (19-0059).
Congratulations to Drs Leran Zhang, Scott Lovell, Elena De Vita, Andrea Goya Grocin and Daniel Lucy and to all our amazing collaborators for the publication of this exciting science!
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Edward Bartlett
Department of Chemistry