Overview
My group uses molecular sequencing for phylogenetic and evolutionary analyses, to establish relationships of living organisms and their genes and genomes. Large-scale DNA analyses are also being used in taxonomy ("DNA taxonomy"), providing diagnostic sequences for each species on Earth. We are applying these DNA approaches to the molecular systematics of insects and specifically the order Coleoptera (beetles). The insects are extremely species rich and include well over half of all known species of animals, providing an enormous challenge for DNA approaches simply due to their great diversity. We are asking, how is the great species richness of organismal lineages distributed on Earth, and what are the evolutionary causes of diversity?
 We conduct most of our work at the Natural History Museum. The research programme links to the broad questions traditionally addressed in Entomology about the taxonomy, phylogenetics and evolutionary biology of insects. For example, we are using a combined bioinformatics and molecular systematics approach to study the deep level phylogeny of the nearly 200 families and major groups of Coleoptera, based on tens of thousands of taxa and large-scale tree searches. Next-generation sequencing has greatly changed this field in the past few years. Our group continually applies latest methods developed for genomics and adapts them to applications in molecular systematics and the study of species diversity.
We conduct most of our work at the Natural History Museum. The research programme links to the broad questions traditionally addressed in Entomology about the taxonomy, phylogenetics and evolutionary biology of insects. For example, we are using a combined bioinformatics and molecular systematics approach to study the deep level phylogeny of the nearly 200 families and major groups of Coleoptera, based on tens of thousands of taxa and large-scale tree searches. Next-generation sequencing has greatly changed this field in the past few years. Our group continually applies latest methods developed for genomics and adapts them to applications in molecular systematics and the study of species diversity.
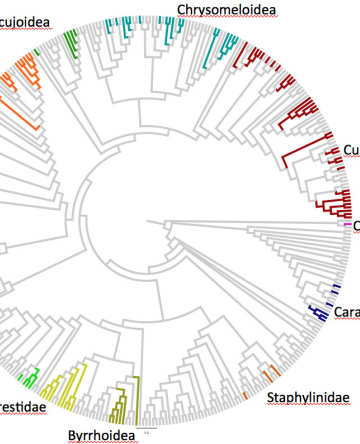 For example, our recent work has used shotgun sequencing mixed specimen samples for rapid and cost-effective generation of whole-mitochondrial genome sequences for phylogenetic tree construction ('mitochondrial metagenomics', MMG). This procedure is equally applied to shotgun sequences of bulk biodiversity samples and efficiently assembles numerous mitogenomes from the mixtures, which are used to place them into phylogenetic trees ('soup-to-tree'). We are currently embarking on a study of such biodiversity samples from around the world, obtaining specimens with standard protocols. Together with collaborators on all continents we use 'site-based' specimen collections to generate a synoptic sample of global species diversity and assemble the tree-of-life, under the umbrella of the SITE-100 project (www.site100.org). This will ultimately provide a better estimate of how many species there are on Earth, a simple but fundamental question for the evolutionary understanding of biodiversity and its conservation. Neutral models of biodiversity provide a useful null model for extrapolations at different hierarchical levels, from clades to species and genotypes.
For example, our recent work has used shotgun sequencing mixed specimen samples for rapid and cost-effective generation of whole-mitochondrial genome sequences for phylogenetic tree construction ('mitochondrial metagenomics', MMG). This procedure is equally applied to shotgun sequences of bulk biodiversity samples and efficiently assembles numerous mitogenomes from the mixtures, which are used to place them into phylogenetic trees ('soup-to-tree'). We are currently embarking on a study of such biodiversity samples from around the world, obtaining specimens with standard protocols. Together with collaborators on all continents we use 'site-based' specimen collections to generate a synoptic sample of global species diversity and assemble the tree-of-life, under the umbrella of the SITE-100 project (www.site100.org). This will ultimately provide a better estimate of how many species there are on Earth, a simple but fundamental question for the evolutionary understanding of biodiversity and its conservation. Neutral models of biodiversity provide a useful null model for extrapolations at different hierarchical levels, from clades to species and genotypes.
Simpler PCR-based 'metabarcoding' protocols are another powerful method for characterising biodiversity samples. The high quality of recent next-gen sequencing now permits accurate species delimitation and numbers in virtually any ecosystem and any organism. We use these methods to study the architecture of insect communities in tropical forests; the spread of tree pathogenic fungi vectored by bark beetles; the transfer of pollen by 'minor' pollinators, especially hoverflies; aquatic freshwater systems in the temperate zone and the tropics. A key  application is the use of this technique to establish trophic interactions and ecological networks, which can be done by deep sequencing of individuals and the detection of 'foreign' DNA obtained by feeding or parasitisation. We thus use individual-based metabarcoding on a large scale with the HiSeq platform.
application is the use of this technique to establish trophic interactions and ecological networks, which can be done by deep sequencing of individuals and the detection of 'foreign' DNA obtained by feeding or parasitisation. We thus use individual-based metabarcoding on a large scale with the HiSeq platform.
Beyond strict biodiversity studies, my group also works on the genetic mechanisms of morphological diversification, using polymorphic swallowtail butterflies as a model. We found that the engrailed locus determines the diversity in female-limited mimetic forms of Papilio dardanus, the African Mocker Swallowtail. Selection on the phenotype has a direct impact on the genome, suppressing recombination and selecting for non-synonymous changes in this region. The ultimate goal is to find the mutations directly responsible for the 'switch' in mimetic phenotype and study its spread across the distributional range. See here for a PhD studentship currently available in this area.
Funding
grants in past 15 years
- The Royal Society: "British and Palestinian museum collaboration in research, education, and conservation". International Collaboration Awards 2019
- DEFRA DNA Centre of Excellence project: "Pollinator-plant interactions and insect community dynamics" 2020
- The Leverhulme Trust: "Genomic and phylogenetic tools for studying species numbers on Earth". International Academic Fellowship 2018
- European Commission: "Twinning for European excellence in Island Biodiversity Genomics (iBioGen)" (NHM/Imperial lead; PI A. Papadopoulou, Univ. Cyprus) 2018
- John Spedan Lewis Foundation. PhD fellowship. 2016
- Royal Society: Newton Advanced Fellowship “Interpreting the evolution of forage selection in small mammals” (Newton Fellow Dr Deyan Ge) 2016
- Biotechnology and Biochemistry Research Council: “Development of the IPI specimen archive as a legacy for future research” 2015
- Department for Environment, Food and Rural Affairs (DEFRA): “Taxonomic fellowship to support the National Pollinator Strategy” 2015
- Natural Environment Research Council: “High-throughput, DNA-based biodiversity assessment and detection for the environmental consultancy market” 2015
- Natural Environment Research Council: “Environmental DNA to test for the presence of endangered species” Impact Acceleration Award 2014
- Royal Society: Newton Programme “Metagenomics to study the unknown diversity of endogeic soil communities” Postdoctoral Fellowship (grant holder Dr Paula Arribas) 2014
- Xunta de Galicia, Spain: Postdoctoral Fellowship (grant holder Dr Carola Gomez-Rodriguez, 2013, (3 years)
- Natural History Museum, Strategic Initiative Fund: “Site-based integrative analysis of biodiversity”, 2012
- Natural Environment Research Council: "Population genomics of invasive fruit flies”, 2012
- Natural Environment Research Council “Female limited Batesian mimicry: epistasis and the path to perfection” (Postdoctoral Fellowship, grant holder Dr Martijn TImmermans), 2011
- Unesco/L’Oreal Women in Science: Postdoctoral Fellowship, grant holder Dr Samia Elfekih, 2011
- Leverhulme Trust: "The beetle Tree-of-Life updated: an expanding resource for taxonomy”, 2009
- Natural Environment Research Council: "Genomic signature of complex morphological traits”
- Natural Environment Research Council: "A DNA resource for phylogenetics and taxonomy of the insects”, 2007
- Natural Environment Research Council: "Correlating wing morphology and genetic variation in mimetic butterflies”, 2006
Research Student supervisIon
- Xueni (Linka) Bian: "Improved phylogenetic models for mitochondrial genomics"
- Sarawut Ounjai: "The phylogenetic diversity of tropical insect communities". Funding: Thai Government fellowship
- Beulah Garner: "The phylogeny and evolutionary ecology of the ground beetle tribe Lebiini". (Part time. Funding: Natural History Museum)
- Chawatat Thanoosing:"The evolution of tropical bumblebees". Funding: Thai Government fellowship
- Angelina Ceballos: “Tracking the associations of tree-pathogenic fungi and insect vectors in space and through time” Funding: John Spedan Lewis Foundation
- Mizanur Rahman: “Biomonitoring of freshwater ecosystems through assessment of pollutants accumulation and biodiversity of invertebrate communities using molecular techniques” (Funding: Commonwealth Fellowship Programme)
previous (year of completion):
- Hannah Norman: "Metagenomics and museum collections to characterise declining pollination webs" (2020)
- Darren Yeo: "Evolution and biogeography of termite inquilines" (2018, co-supervision with Natl. Univ. Singapore)
- Maria Belen Arias: "Population genomics of the medfly, Ceratitis capitata" (2018)
- Thomas Creedy: "Monitoring climate change in tropical forests: building a 21st century tool kit" (2017)
- Kirsten Miller (2015) “DNA profiling of insect herbivory in tropical forests“.
- Alexandra Crampton-Platt (2015) “The structural diversity of insect communities in tropical and temperate forest landscapes“
- Amrita Srivathsan (2015) “Phylogenomics of feeding preferences in the endangered banded leaf monkey”
- Martin Thompson (2014) “The origin and establishment of morphological novelty in the evolution of butterfly mimicry forms“
- Conrad Gillet (2014) “A phylogeny for the Curculionidae: an integrated molecular approach to utilise the biodiversity of museums"
- Chris Barton (2014) “Exploiting DNA databases for species delimitation and intra-specific evolution“
- Douglas Chester (2011) “Molecular evolution and phylogenetic information content of DNA barcodes”
- Tomochika Fujisawa (2011) “Statistical analysis on genealogical-phylogenetic data”
- Benjamin Isambert (2010) “The causes of insect endemicity in biodiversity hotspots”
- Chris Borrow (2010) “Building and testing a global DNA barcode database of dung beetle assemblages”
- Sara Pinzon (2010) “Evolutionary ecology of weevil seed predation in tropical forests”
- Anna Papadopoulou (2009) “Phylogeography of tenebrionid beetles in the Aegean islands”
- Toby Hunt (2007) “Evolutionary bioinformatics of Coleoptera”
- Rebecca Clark (2006) “Evolution of wing patterns in the Mocker Swallowtail, Papilio dardanus”
- Stuart Longhorn (2005) “Phylogenomics of Coleoptera”
- Joseph Hughes (2004) “The evolution and ecology of seed predation in weevils (genus Curculio)”
- Claire Wilsher (2003) “Ecomorphological and phylogenetic analysis of dung beetle communities”
- Daegan Inward (2003) “The evolution of dung beetle assemblages”
- Jason Mate (2003) “Radiation and diversification of Aphodius dung beetles”.
- Tom Anthony (2003) “Vector partitioning and mating patterns in Plasmodium falciparum”
- Harriet Green (2001, not awarded) “The molecular evolution of sexual and asexual lineages in the bark beetle Ips acuminatus”
- Gail Reeves (2001) “Radiation and macroevolutionary ecology of the African genus Protea”.
- Karen Miller (2000) "Evolution of retroviruses"
POSTDOCTORAL FELLOWS
- Dr Huaxi Liu
Previous:
- Dr Thomas Creedy (2018-2021)
- Dr Deyan Ge (2016-2020)
- Dr RuiE Nie (2017-2018)
- Dr Alejandro Lopez (2016)
- Dr Pu Tang (2015 – 2016)
- Dr Cuong Tang (2015 – 2016)
- Dr Paula Arribas (J2014 – 2016)
- Dr Carola Gomez-Rodriguez (2013 – 2016)
- Dr Carmelo Andujar (2013 – 2016)
- Dr Benjamin Linard (2013 – 2016)
- Dr Martijn Timmermans (2008 – 2014)
- Dr Debora Pires Paula (2013 – 2014)
- Dr Samia Elfekih (2011 – 2013)
- Dr Sujatha N. Kutty, (2011 – 2013)
- Dr Cesc Murria i Farnos (2009 – 2011)
- Dr Johannes Bergsten (2005 – 2009)
- Dr Michael Monaghan (2003 – 2008)
- Dr Toby Hunt (2007)
- Dr Raul Bonal (2006 – 2008)
- Dr David Lees (2003 – 2007)
- Dr Rebecca Clark (2006 – 2007)
- Dr Chao-Dong Zhu (2004 – 2006)
- Dr Joan Pons (2001 – 2005)
- Dr Michael Balke (2002 – 2005)
- Dr Ryan Gregory (2003 – 2004)
- Dr Joseph Hughes (2003 – 2004)
- Dr Ruxandra Bucur (2004)
- Dr Richard Davies (2001– 2003)
- Dr Jesus Gomes-Zurita (2000 – 2003)
- Dr Kosmas Theodoridis (2001 – 2004)
- Dr Miquel Arnedo (2002)
- Dr Michael Caterino (1999 – 2001)
- Dr Ignacio Ribera (1998 – 2002)
- Dr Alexandra Cieslak (1998 – 2002)
- Dr Anthony Cognato (1999 – 2000)
- Dr Tim Barraclough (1996 – 1999)
- Dr Verel Shull (1996 – 2000)
-
MEMBERSHIPS
- Research Assessment Exercise, University of Oulu, Finland, 2021
- DNAqua-Net, H2020 COST Action CA15219, UK representative and member of Management Committee, 2016 - 2021
- NatureMetrics Ltd., Egham, Surrey, UK; Co-founder and member of Scientific Advisory Board, 2015 - present
- University College London, Genetics, Evolution & Environment; Honorary Professor, 2013
- Chinese Academy of Sciences, Institute of Zoology, Visiting Professor, 2009
- Research Council of Norway, grant review panels ‘Young Outstanding Investigators’ (2007) and ‘Independent Projects – Biology and Biomedicine (FRIBIO)’ (2007-2009)
- International Barcode-of-Life Initiative (iBOL), UK Representative (2009-2011)
- Systematics Association UK, Member of Council (2007-2009)
- NERC Peer Review College, Member (2006-2010)
- NERC Molecular Genetics Facilities, Member of Management Committee (2004-2009)
- German Research Association (DFG), Peer Review Committee, Priority Programme ‘Adaptive Radiation’, (2003-2008), Priority Programme ‘Deep Metazoan Phylogeny’ (2004-2009)
- Society for Systematic Biology, Member of Council (2000-2003)
Editorial Boards
- Associate Editor, Molecular Phylogenetics and Evolution (2008-present)
- Editorial Board, Journal of Zoological Systematics & Evolutionary Research (2006-present)
- Advisory Council, Zoologica Scripta (2005-present)
- Editorial Board, BMC Evolutionary Biology (2005-present)
- Associate Editor, Conservation Genetics (2002-present)
- Editorial Board, Journal of Evolutionary Biology (2002-2007)
- Editorial Board, Systematic Entomology (1998-2008)

