Overview
We work on signalling systems in plant defence and development, that allow plants to function in an integrated manner. The greatest present challenges are to identify novel regulatory molecules and their targets. Fortunately, functional genomics enables us to address these questions directly. In particular, we can exploit proteomics and transcriptomics in combination with the widespread availability of mutants and transgenics. For plant defence against aphids, we are particularly interested in species-wide diversity of both pest virulence and host resistance (Kanvil et al. 2014) and in genetics of aphid effectors (Kanvil et al. 2015). We have also invested time in developing tools for studying systemic signalling in Arabidopsis, especially micro-grafting and sap sampling. We have published on both phloem (Corbesier et al. 2007; Truman et al. 2007; Zhang et al. 2010) and xylem (Foo et al. 2007) systems.
Major research activities
- Molecular basis of virulence and avirulence in aphids, and their immune-suppressing or immune-activating functions in host plants
- Hormonal control of shoot architecture (branching) and developmental switches (flowering, dormancy)
- Long-distance transmission of macromolecular signals in phloem
- Structure and function of proteins controlling flowering time
Current research projects
1. Molecular Biology of aphid virulence and host plant resistance
BBSRC project 2022-2025: Functions of a novel chitinase-like effector family unique to aphids
Virulent pea aphid feeding on Medicago host
Aphids are major worldwide crop pests that cause damage by feeding on phloem sap and frequently by acting as vectors for transmission of numerous viruses. Although some aphid resistance genes have been identified in host plants, very little is known of aphid genetics in relation to why some genotypes are highly virulent on particular hosts and not others. Aphids deliver effector molecules into the host, in an analogous manner to effector delivery from bacterial, fungal and oomycete pathogens. But we know little of aphid effector targets in host plants that result in immune suppression or metabolic reprogramming. Equally, we do not yet know the pathways that lead to effective resistance in presence of appropriate R-genes: what kills or deters aphids? We have screened for candidate effectors by transcriptomics and proteomics, leading to identification of a novel class of proteins with weak homologies to chitinases, that we call CHitinase-Like (CHL), that are abundant in saliva but completely uncharacterised. These proteins are unique to aphids, and may provide clues to their global success as phloem-feeders. Our new BBSRC-funded project is exploring the biochemical functions of CHL proteins, and examining mechanisms of host immune suppression by aphids.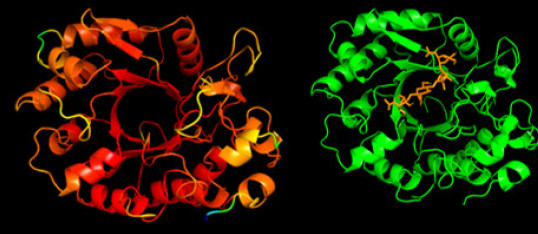
AlphaFold model of CHL1 protein, with predicted docking of chitin oligomer ligand
2. Regulation of meristem dormancy and shoot branching architecture
We aim to understand how systemic signalling regulates shoot architecture, and then to examine possibilities for novel means of regulating the extent of branching. Potato tuber bud dormancy is regulated by similar signals and we have recently been addressing the major industry problem of sprouting in storage. There are many further instances in agriculture, horticulture and forestry where yield, quality and net value of crop species are closely linked to stem numbers.
Shoot branching model predict multiple hormone signals, some of which are systemically mobile via xylem or phloem. One of these was identified as strigolactone (Gomez-Roldan et al., Nature, 2008), derived from carotenoid cleavage pathways. We are attempting to define the molecular and biochemical regulatory relationships between strigolactones and the traditionally recognised branching regulators, cytokinin (Foo et al. 2007; Young et al. 2014) and auxin. Strigolactone defects (max mutants) and cytokinin deficiency (ipt knockouts or CKX overexpression) have very different and additive effects on phenotype (see image above). We have also been examining cytokinin distribution and perception at the cellular level using cell sorting in conjunction with high sensitivity mass spectrometry (Antoniadi et al. 2015, 2020).
3. Molecular biology of graft transmissible flowering signals
Florigen(s) - mobile substances that control flowering - have proved elusive for decades. But we now know that macromolecules - mRNA and/or proteins - rather than small molecule hormones may be responsible. Using our Arabidopsis grafting methods and confocal microscopy, we discovered that biologically active GFP fusions to a key photoperiod pathway protein, FT, can be transmitted through the phloem towards the shoot apex. The images here show graft transmission of FT:GFP into shoot tip (left) and root tip (middle, right). Based on this and other experimental evidence, we proposed that FT protein itself is the active and mobile florigen hormone (Corbesier et al. 2007). We are now using proteomics and biochemistry to screen for additional components of the signalling complex, and to search for new ways to reliably manipulate flowering time.
CURRENT VACANCIES
Interested in INDEPENDENT Postdoctoral Fellowships?
Imperial College runs an annual competitive Imperial College Research Fellowships scheme (Imperial College Research Fellowships). If you are eligible and might be interested in applying, please get in touch at c.turnbull@imperial.ac.uk. We can also assist in preparing applications for other indepedent fellowship schemes run by Royal Society, BBSRC and many other organisations.
Interested in PhD or MRes projects?
If you are interested in projects within our research areas on plant defence or development, please get in touch at c.turnbull@imperial.ac.uk.
The lab currently is focussed on plant-aphid interactions: inheritance and characterisation of effectors and resistance genes, mechanisms of immune activation and immune suppression, and climate change impacts on plant immunity.
Publications
Selected Research Papers
Antoniadi I, Novák O, Gelová Z et al., 2020, Cell-surface receptors enable perception of extracellular cytokinins. Nature Communications 11: 4284. Link
Kanvil S, Pham J, Lopez‐Cobollo R et al., 2017, Cucurbit extrafascicular phloem has strong negative impacts on aphids and is not a preferred feeding site. Plant, Cell & Environment 40: 2780-2789 Link
et al., 2016, Comparative proteomics of cucurbit phloem indicates both unique and shared sets of proteins. Plant J. 88: 633-647 Link
et al., 2015, Cell-Type-Specific Cytokinin Distribution within the Arabidopsis Primary Root Apex, Plant Cell, 27, 1955-1967 Link
Kanvil S, Collins CM, Powell G, Turnbull CGN. (2015) Cryptic virulence and avirulence alleles revealed by controlled sexual recombination in pea aphids. Genetics 199: 581-593. Genetics Abstract.
Kanvil K Powell G Turnbull C (2014) Pea aphid biotype performance on diverse Medicago host genotypes indicates highly specific virulence and resistance functions. Bulletin of Entomological Research 6: 689-701. doi.org/10.1017/S0007485314000443
Young NF Ferguson BJ, Antoniadi I, Bennett MH, Beveridge CA, Turnbull CGN (2014) Conditional auxin response and differential cytokinin profiles in shoot branching mutants. Plant Physiology 165:1723-1736. plantphysiol.org/content/165/4/1723.abstract
Bromley JR, Warnes BJ, Newell CA, Thomson JC, James CM, Turnbull CGN, Hanke DE (2014) A purine nucleoside phosphorylase in Solanum tuberosum L. (potato) with specificity for cytokinins contributes to the duration of tuber endodormancy. Biochemical J. 458, 225-37.biochemj.org/bj/458/bj4580225.htm
Turnbull CGN, Lopez-Cobollo RM (2013) Tansley Review: Heavy traffic in the fast lane: long-distance signalling by macromolecules., New Phytol, Vol:198, Pages:33-51 http://onlinelibrary.wiley.com/doi/10.1111/nph.12167/abstract
Braun N, de Saint Germain A, Pillot JP, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching., Plant Physiol, 2012, Vol:158, Pages:225-238 www.plantphysiol.org/content/158/1/225 (doi)
Turnbull C (2011) Long-distance regulation of flowering time. J Exp Bot, 62, 4399-4413 http://jxb.oxfordjournals.org/content/62/13/4399 (doi)
Zhang B, Tolstikov V, Turnbull C, Hicks LM, Fiehn O (2010) Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proceedings of the National Academy of Sciences USA 107, 13532-13537. http://www.pnas.org/content/early/2010/06/10/...
Truman WM, Bennett MH, Turnbull CGN, Grant MR (2010) Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiology 152, 1562-1573. www.plantphysiol.org/cgi/content/abstract/152/3/1562
Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21, 39-53. www.plantcell.org/cgi/content/abstract/21/1/39
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signalling in floral induction of Arabidopsis, Science 316, 1030 - 1033. www.sciencemag.org/cgi/content/abstract/316/5827/1030
Foo E, Morris SE, Parmenter K, Yo ung N, Wang H, Jones A, Rameau C, Turnbull CGN, Beveridge CA (2007) Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiology, 143, 1418-1428 www.plantphysiol.org/cgi/content/abstract/143/3/1418
Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. ( 2007) Arabidopsis systemic immunity uses conserved defense signalling pathwa ys and is mediated by jasmonates. Proceedings of the National Academy of Sciences USA 104, 1075-1080. www.pnas.org/cgi/content/abstract/104/3/1075
Dodd IC, Ngo C, Turnbull CGN, Beveridge CA (2004) Effects of nitrogen supply on xylem cytokinin delivery, transpiration and leaf expansion of pea genotypes differing in xylem cytokinin concentration. Functional Plant Biology 31, 903-911.
An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615-3626. http://dev.biologists.org/cgi/content/abstract/131/15/3615
Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant Journal 32, 255-262.
Morris SE, Turnbull CGN, Murfet IC, Beveridge CA (2001) Mutational analysis of branching in pea (Pisum sativum L.): Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiology 126, 1205-1213.
Foo E, Turnbull, CGN, Beveridge, CA (2001) Long-distance signaling and the contr ol of branching in the rms1 mutant of pea. Plant Physiology 126, 203-209.
Beveridge CA, Symons GM, Turnbull CGN (2000) Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiology 123, 689-696.
Turnbull CGN, Raymond MAA, Dodd IC, Morris SE (1997) Rapid increases in cytokinin concentration in lateral buds of chickpea (Cicer arietinum L.) during release of apical dominance. Planta 202, 271-276.
Books/Book Chapters
Turnbull CGN (2014) Grafting in Arabidopsis. In: Arabidopsis Protocols3rd edition, Methods in Molecular Biology 1062, Sanchez-Serrano JJ, Salinas J, eds, (Humana Press) ISBN 978-1-62703-579-8
Turnbull CGN (2010) Grafting as a Research Tool. In :Plant Developmental Biology, Methods in Molecular Biology 655, Hennig L, Köhler C eds., DOI 10.1007/978-1-60761-765-5_2. (Springer Science Business Media) ISBN 978-1-60761-764-8
Turnbull CGN (Ed) (2005) Plant Architecture and Its Manipulation. Annual Plant Reviews Volume 17. (Blackwell Publishing, Oxford) ISBN 1-4051-2128-9/ISSN 1460-1494
Bainbridge K, Bennett T, Turnbull C, Leyser O (2006) Grafting. In: Arabidopsis Protocols, 2nd edition, Salinas J, Sanchez-Serrano JJ, eds, (Humana Press, Totowa, USA). ISBN 1-58829-395-5.
Atwell, BJ, Kriedemann, PE, Turnbull, CGN (Eds) (1999) Plants in Action: Adaptation in Nature, Performance in Cultivation. (Macmillan: Melbourne). ISBN 0-7329-4439-2.
Research Staff
Kanvil,S
Lopez-Cobollo,R
Research Student Supervision
Antoniaidi,I, Regulation and function of the plant hormone cytokinin as a long distance signal
Joyce,W, Chemical Biology of Flowering Time Control
patel,S, Chemical proteomics and fatty acid elongation

