RESEARCH
 Protein folding and CCT
Protein folding and CCT
The cytosolic chaperonin CCT is a 1-MDa multi-subunit protein complex that has an essential, core function in actin protein folding in all eukaryotes. In 2011 we determined the atomic structure of the closed form of this yeast CCT-actin complex by X-ray crystallography. The structural studies all point to a remarkably asymmetrical machine with the features required to support a sequential allosteric folding mechanism able to bind specifically and anneal the non-native actin polypeptide. We believe that the final outcome of the CCT-dependent actin folding mechanism is to produce a protein spring whereby the actin monomer is able to reversibly re-explore the folding landscape in the structural context of the polymeric F-actin filament. This model has been extended and refined in my recent review article: Biochemical Journal (2018) 475 3009–3034 - The structure and evolution of eukaryotic chaperonin-containing TCP-1 and its mechanism that folds actin into a protein spring.
You can listen to a 40 minute recorded talk: Willison, K. R. (2021). Chaperonin-containing TCP-1 (CCT), actin springs, and protein folding fluxes. In The Biomedical & Life Sciences Collection, Henry Stewart Talks. Retrieved June 16, 2021, from https://hstalks.com/bs/341/.
Malarial CCT

Mark Wilkinson, was a 4-year EPSRC Chemical Biology CDT PhD student (2015-2020). Targeting protein folding in the malaria parasite (PhD awarded 2020) with Dr Jake Baum (Life Sciences), Prof Keith Willison (Chemistry).
Truncated latrunculins as actin inhibitors targeting plasmodium falciparum motility and host cell invasion (2016) Journal of Medicinal Chemistry DOI:10.1021/acs.jmedchem.6b01109
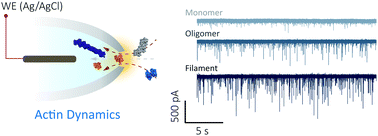
Single-molecule nanopore sensing of actin dynamics and drug binding (2020) Chemical science DOI:10.1039/C9SC05710B
A biosynthetic platform for antimalarial drug discovery (2020) Antimicrobial Agents and Chemotherapy DOI:10.1128/AAC.02129-19
SINGLE CELL PROTEOMICS

Single molecule counting approaches are not only an essential approach for clinical and pre-clinical science and systems medicine but also for generating precise quantitative data for the mathematization of biology. The holy grail of single cell proteomics is the ability to count, within high dynamic range, the copy numbers, protein-protein interactions and post-translational modifications of many proteins in individual cells using label-free approaches. Here, label-free means that the proteins are not pre-labelled before analysis: for example by using GFP-gene tagging or in vivo chemical labelling technologies. Prof David Klug and I are working to develop robust, high-throughput methods to count, at single molecule sensitivity, proteins found in bodily fluids and in rare cells present in clinical biopsies. Single molecule detection is highly accurate, does not require calibration and the read-out is digital. We have created a multi-disciplinary laboratory environment to implement and to invent the tools and technologies (T&Ts) needed to achieve these ambitious goals. At this stage in the development of this field most of the T&Ts are by no means plug and play and require expertise in engineering (microfluidics), applied optics (total internal reflection fluorescence and optical trapping spectroscopies), mathematics (data analysis and model building), cell biology (cell separation and manipulation), protein chemistry (antibody development) and chemistry (probe use and development). The first prototype device is the MAC chip (Microfluidic Antibody Capture) which has been used to count p53 tumor suppressor proteins at single molecule level in single colorectal cancer cells. Willison,K.R. and Klug, D.R. (2013) Current Opinion in Biotechnology 24, 745-751.
NANOTECHNOLOGY

We are working with the group of Prof Joshua Edel on various aspects of nanoprobes applied to problems in cell biology and protein dynamics. Our first study on single-cell biopsies was published in Nature Nanotechnology in 2019.
Press release: https://www.imperial.ac.uk/news/189227/nanoscale-tweezers-perform-single-molecule-biopsies-individual/
In 2020 we published a nanopore analysis of actin dynamics and drug binding at single-molecule resolution. We have been able to follow actin unfolding kinetics and F-actin polymerization in a label-free configuration thus opening up fundamentally new approaches to probe the energy landscape of this essential eukaryotic protein.
Single-molecule nanopore sensing of actin dynamics and drug binding
Xiaoyi Wang, Mark D. Wilkinson, Xiaoyan Lin, Ren Ren, Keith R. Willison, Aleksandar P. Ivanov, Jake Baum and Joshua B. Edel
Nanopipettes were used for real-time investigation into actin dynamics and drug binding at single-molecule resolution, showing promise for a better understanding of the mechanism of protein–protein interactions and drug discovery.
Protein Aggregation

Abeta(40) and α-Synuclein
We have worked with Dr Liming Ying's group on protein aggregation processes concerning the peptides Abeta(40) and α-Synuclein which are involved in human neurodegenerative disease states.
Acetylation Rather than H50Q Mutation Impacts the Kinetics of Cu(II) Binding to α-Synuclein.
Teng X, Sheveleva A, Tuna F, Willison KR, Ying L. Chemphyschem (December 2021)
Guest Lectures
Willison, K. R. (2021). Chaperonin-containing TCP-1 (CCT), actin springs, and protein folding fluxes [Video file]., In The Biomedical & Life Sciences Collection, Henry Stewart Talks., Henry Stewart Talks, Ruskin House, 40/41 Museum StreetLondon WC1A 1LT, 2021
Thinking about CCT - evolution and the actin folding mechanism, Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel, 2019
An intracellular calcium wavelength code model extended to the Riemann zeta function, EPSRC Physics of Biological Oscillators: new insights into non-equilibrium and non-autonomous systems, RS Chicheley Hall, Buckinghamshire, UK, 2018
Counting single protein molecules in rare single cells isolated from human clinical samples, Graduate School of Advanced Sciences of Matter, Hiroshima University, Japan, 2017
Counting single protein molecules in rare single cells isolated from human clinical samples, Department of Biotechnology and Life Science Tokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, Tokyo Japan 184-8588, 2017
Counting single protein molecules in rare single cells isolated from human clinical samples, Tokyo Institute of Technology, Cell Biology Center, Institute of Innovative Research, Suzukakedai Campus, Tokyo, Japan, 2017
Quantitative single cell proteomics and theoretical approaches to cytoplasmic organization and neural codes using non-continuous zeta functions, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, Japan, 2016
Quantitative single cell and single molecule proteomics for clinical studies, National Taiwan University (NTU)Graduate Institute of Biomedical Electronics and Bioinformatics, Taipei, Taiwan, 2016
Quantitative single cell and single molecule proteomics for clinical studies, National Tsing Hua Uniersity (NHTU), Hsinchu, Taiwan, 2016
Quantitative single cell and single molecule proteomics for clinical studies, National Health Research Insitiutes (NHRI)Institute of Biomedical Engineering and Nanaomedicine, Hsinchu, Taiwan, 2016
Absolute quantification of protein copy number in single cells using single molecule microarrays, 10th European Biophysics Congress European Biophysics Journal Vol 44. Supplement 1. July 2015. S179. O-524, International Congress Center Dresden, Germany, 2015
Quantitative Single Cell and Single Molecule Proteomics for Clinical Studies, CHI Precision Diagnostics Summit, Cambridge Healthtech Institute, 250 First Avenue, Suite 380, Needham, MA 02494, USA, 2014
Eukaryotic cytosolic chaperonin CCT: annealing actin into a spring, Ecole Polytechnique Federale De Lausanne and Universite de Lausanne, 25 years of chaperone research: Protein Folding, in and out of Anfinsen's closet. Symposium held in Arolla, Switzerland, 2014
Quantitative single cell and single molecule proteomics for clinical studies, Institute for Virus Research, Kyoto University, Kyoto, Japan, 2013
Eukaryotic cytosolic chaperonin CCT: the actin folding mechanism and cell biology in yeast, Graduate School and Faculty of Engineering and Resource Science, Akita University, Akita, Japan, 2013
Eukaryotic cytosolic chaperonin CCT: the actin folding mechanism produces a protein spring, Department of Biotechnology and Life ScienceTokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, TokyoJapan 184-8588, 2013
Eukaryotic cytosolic chaperonin CCT: the actin folding mechanism and the CCT-interactome, Department of Materials Science and Chemistry, Graduate School of Engineering, University of Hyogo, Himeji, 671-2280 Japan, 2013
Eukaryotic cytosolic chaperonin CCT: the actin folding mechanism and the CCT-interactome, Israel Science Foundation Workshop - Protein folding: moving beyond simple model systems, Weizmann Institute of Science, Rehovot, Israel, 2012
The eukaryotic chaperonin CCT: mass spectrometric analysis of CCT binding proteins, Molecular Chaperone Club Annual Meeting, Department of Structural Biology, Birkbeck College, University of London, London, 2011
Structure-function analysis of the eukaryotic cytosolic chaperonin CCT: folding actin uphill into a spring-like state, CIHR Training Grant in Protein Folding and Interaction Dynamics, University of Toronto, University of Toronto, Toronto, Canada, 2011
Analysis of the evolution of the eukaryotic CCT-actin folding system using structural and network biology data, Center for Comparative Genomics and Evolutionary Bioinformatics, Dalhousie University, Dalhousie University, Halifax, Nova Scotia, Canada, 2011
The actin and tubulin folding CCT ATPase: an allosteric protein machine as a potential cancer drug target, Pharmazeutischen Institute, Technische Universitat Braunschweig, Braunschweig, Germany, 2010
The eukaryotic chaperonin CCT:structure/function analysis of its core activity in folding actin and some WD40 repeat-containing proteins., ZOMES VI Conference, Safed, Safed, Israel, 2010
Crystal structure and functional studies of the eukaryotic chaperonin CCT, Biophysics Program, Yale University, Newhaven, CT, USA, 2009
The eukaryotic chaperonin CCT: structure, function and systems analysis of its core activity in folding actin, University of Virginia, Charlottesville, Virginia, USA, 2009
Structure of the eukaryotic chaperonin CCT, Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel, 2009
The eukaryotic chaperonin CCT: structure, function and systems analysis of its core activity in folding actin, Safed Workshop on Biomolecular modeling and simulations, Safed, Israel, 2009
Constructing the functional interaction network of a protein folding machine-the CCT-actin system, Lundberg Laboratory Goteborgs University, Goteborgs, Sweden, 2009
Constructing the interaction network of a protein folding machine-the CCT-actin system, Institute of Cell and Molecular Science, Queen Mary's College, QMC, University of London, 2008
The eukaryotic chaperonin CCT: its interaction network and functional analysis of its actin folding activity, Brigham Young University, Salt Lake City, Utah, USA, 2008

