Projects
Current and previous projects are described below.
power analysis in metabolomics
We have developed a new tool for estimating power and sample size for metabolomics studies. The approach is based on realistic simulations using pilot datasets, and captures the correlation and heavy tailed characteristics of typical metabolomic data. Work in collaboration with Benjamin Blaise and Goncalo Correia. See the paper. You can run the tool on the Phenomenal Galaxy site (see 'STATS').
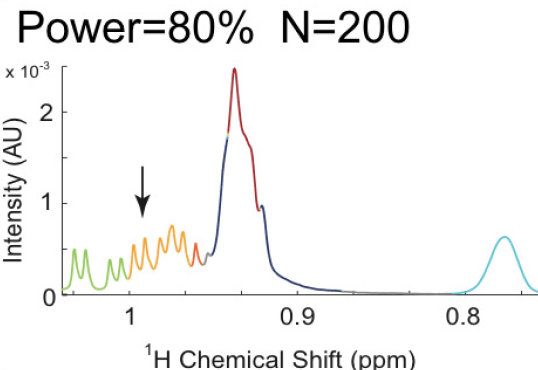
BATMAN: BAYESIAN ANALYSIS OF METABOLIC NMR SPECTRA
In this collaboration with Dr Maria De Iorio, UCL, we have developed a Bayesian model of NMR spectra which aims to automatically deconvolve and quantify resonances of metabolites in complex 1-dimensional spectra of biofluids and tissues. We have released a publicly available R package called the Bayesian AuTomated Metabolite Analyser for NMR spectra (BATMAN).
BATMAN has an R-Forge site where you can download the package. Alternatively you can install direct from R by typing:
install.packages("batman", repos="http://R-Forge.R-project.org").
You can also read the papers in Bioinformatics, Journal of the American Statisitical Association and Nature Protocols.
SIMULATION OF NMR METABOLIC PROFILES
With Dr Maria De Iorio, now at UCL, we developed a MATLAB program which is able to simulate realistic 1-dimensional 1H NMR spectra of complex mixtures of metabolites. The programme 'MetAssimulo' allows the user to specify the levels and variability of metabolites to be incorporated, and by default will simulate two groups corresponding to 'cases' and 'controls'. MetAssimulo can also simulate correlations within and between metabolites, as well as shifts in peak positions often seen in metabolic spectra. The picture below shows two NMR spectra of human urine. One is real and one is simulated. Can you tell which is which?
For more information please visit the MetAssimulo page or read the paper.
TIME SERIES ANALYSIS IN METABOLOMICS
With Giovanni Montana (now at Kings) we have developed new techniques for time series analysis in metabolomics. Traditional time series methods require hundreds of time points and typically monitor just a few variables. In metabolic profiling, we monitor hundreds to thousands of variables over just a few (typically 3-10) time points. The plot below shows an example from our smoothing splines mixed effects (SME) model where the time course for two different groups of animals is being compared for a single metabolite. See publication.
PATHWAY TOOLS FOR INTEGRATING MULTI-OMICS DATA
With Hector Keun, Rachel Cavill (Imperial) and Ralf Herwig and Atans Kamburov (MPIMG, Germany) we developed new tools which allow researchers to interpret data from multiple omics experiments in a pathway context. The pathway approach allows a higher sensitivity and more global overview of the effects in a biological study and is particularly suited to integrating data from different levels of biomolecular organisation (e.g. metabolomics, transcriptomics) obtained in heterogeneous experiments. The figure blow shows a clustering of 118 drugs (rows) with hundreds of pathways (columns) indicating the overall pathway response of different cancer cell lines to drug treatment, as measured by metabolomics and transcriptomics. See the paper and the online tool IMPaLA.
DIFFERENTIAL CORRELATION NETWORKS
A characteristic of omics data is that the individual measurements are highly correlated. That is, the level of a gene or metabolite tends to vary in a similar way to that of other genes/metabolites. These correlations can be visualised as a network in which correlated molecules are connected. The correlation network can be thought of as a new type of 'fingerprint' of the biological status of an organism and is therefore of interest in its own right. With Maria De Iorio (UCL) we have developed methods for analysing how correlation networks change in response to different biological conditions. We term these 'differential correlation networks'. The picture below shows which links in the blood lipoprotein correlation network are different between normal people and those with pre-diabetics symptoms. See the paper1, paper2 and paper3.
CLASSIFICATION TOOLS FOR METABOLOMICS
COMET Expert System: This figure shows a schematic of an 'expert system' constructed to help predict the toxicity of novel drugs. On the left NMR spectra of rat urine are shown (a,b), a model of normality is developed (c) over a time course (d). The right panel shows a similarity matrix comparing metabolic profiles from 62 treatments affecting the liver or kidney reflecting a high degree of organ specificity. See Ebbels et al. J. Prot. Res. 2007.
RIGOROUS STATISTICAL MODELS OF LIQUID CHROMATOGRAPHY MASS SPECTROMETRY (LC-MS) DATA
LC-MS is one of the most widely used technologies in metabolomics and other bioanalytical sciences today. The measurement process is very complex and thus most methods for processing the data rely on heuristic approaches. With Andreas Ipsen we have developed a new model of LC-Time of Flight-MS, based on the physical ion counting process at the detector, which is able to model the data more accurately than ever before. The model has so far led to two applications: detection of co-eluting compounds and construction of rigorous confidence intervals on isotope ratios. Both of these applications help in the difficult task of identifying unknown molecules - one of the key bottlenecks in metabolomics today. The picture below shows the ion counts from two partially co-eluting species. The colours display the p-value from the new test indicating clearly that these two peaks do not co-elute. More details in the papers 1, 2, 3 & 4
Collaborators
MRC-HPA Centre for environment and Health, Imperial College London, 2010
Dr Maria De Iorio, University College London, Bayesian Biostatistics
Guest Lectures
Pathway space transformation for metabolomics focussed integration of multi-omics data, Metabolomics in Life Science Workshop, University of Umea, Sweden, 2024
Multi-block chemometrics and pathway scoring for interpretation and integration of metabolomics and multi-omics data, Syngenta Informatics Webinar Series, online, 2023
Metabolomics driven data integration using biological pathways, University of Georgia Bioinformatics Seminar Series, online, 2023
Pathway Analysis in Metabolomics, SymbNET Workshop, Lausanne, Switzerland, 2023
Computational analysis: the crucial ingredient in modern metabolomics, INRAE, Toulouse, France, 2023
Problems, principles and progress in computational annotation of NMR metabolomics data, Metabolomics Journal Hot Topics Workshop, online, 2023
Metabolism Meets Machine Learning: Computational Metabolomics, Professorial Inaugural Lecture, Imperial College London, 2023
Computational Metabolomics in Biomedical Research, Dept Metabolism, Digestion & Reproduction, Imperial College, Kensington, London, 2023
Interpreting and Integrating Metabolomics Data with Pathway Methods, Harvard Metabolomic Epidemiology Seminar, online, 2022
Computational Metabolomics: Integration and Interpretation using Pathways, LAMPS Latin American Metabolic Profiling Society, Cartagena, Columbia, 2022
Pathway methods in metabolomics: Integration & Interpretation, Frontiers in Metabolomics Workshop, Telluride, Colorado, USA, 2022
Data Science Challenges and Reporting Needs in Metabolomic Epidemiology, Metabolomics2022, Conference of the Metabolomics Society, Valencia, Spain, 2022
Metabolomics-driven data integration: challenges and solutions, XOmics Festival, Nijmegen, The Netherlands, 2022
Processing and Analysis of Metabolomics Data, European School of Metabolomics, Moustiers-Sainte-Marie, France, 2022
Modelling Complex Metabolomic NMR Spectra with BATMAN, Royal Society of Chemistry Molecular Spectroscopy Group webinar series, online, 2022
Computational Metabolomics: molecules meet machine learning, Sargent Centre Webinar, Imperial College London, online, 2022
Computational Metabolomics (& other omics stories), ViBo Quasar Meeting, University of Liverpool, online, 2022
Computational Metabolomics: Molecules Meet Machine Learning, Imperial College London, London, 2021
Computational Metabolomics: what is it and why do we need it?, Metabolomics Society Early Career Members Network, webinar, 2021
The Data Science Helper Team on the MRes in Biomedical Research, Talking Teaching, Imperial College London, webinar, 2021
Towards Solving the Grand Challenges of Computational Metabolomics, UK Conference on Bioinformatics and Computational Biology, Online, 2020
Computational Metabolomics, Dagstuhl Workshop on Computational Metabolomics, Dagstuhl, Germany, 2020
Computational Metabolomics: The Key to the Future?, Scottish Metabolomics Network Conference, Glasgow, UK, 2019
Sparse Multiblock PLS for Selection of Related Signals in Multi-platform Metabolomics Data, Metabolomics 2019 Conference, The Hague, Netherlands, 2019
Data Integration in Metabolomics: Concepts and Examples, Metabolomics 2019 Conference, The Hague, Netherlands, 2019
Challenges and Solutions in Analysing Large Scale Metabolomics Data, University of Umea, Umea, Sweden, 2019
Challenges in Processing and Integrating Multi-cohort Metabolomics Data, Health Data Research UK, Kings College London, 2019
Challenges in Processing and Analysing Large Scale LC-MS Metabolomics Data, UK Biobank, UK Biobank Metabolomics Workshop, Wellcome Building, London, 2018
Statistical Tools for Data Integration in Metabolomics, Telluride Metabolomics Workshop, Telluride, CO, USA, 2018
Integrating Metabolomics and Other Omics Data: Chemometric and Network Tools, Netherlands Conference on Systems Biology & Bioinformatics BioSB, Congrescentrum De Werelt, Lunteren, Netherlands, 2018
Metabolomics in the Cloud: Scaling Computational Tools to Big Data, Phenomenal Project Webinar Series, Webinar, 2018
Metabolomics in the Cloud: Scaling Computational Tools to Big Data, Metabomeeting 2017, University of Birmingham, Birmingham, UK, 2017
Data Analytical Challenges in Large Scale Metabolomics, University of Georgia, Athens, Georgia, USA, 2017
Computational Metabolomics, Shanghai Jiaotong University, Shanghai, China, 2017
Computational Metabolomics, Zhejiang University Children’s Hospital, Zhejiang, China, 2017
Strategies for data integration in omics: a metabolomics viewpoint, Dupont ATOMS Seminars, Dupont ATOMS webinar series, 2017
Data Analytical Challenges in Large Scale Metabolomics, Euroanalysis Conference, Stockholm, Sweden, 2017
Power analysis and sample size determination in metabolic phenotyping, Workflow4Experimenters Workshop, Paris, 2017
Computational Metabolomics at Imperial College London, Shonan Meeting on Computational Metabolomics, Tokyo, Japan, 2017
Power analysis and sample size determination in metabolic phenotyping, SMPGD Conference, Imperial College London, 2017
Bayesian Deconvolution and Quantitation of 1-D NMR Metabolic Profiles with BATMAN, University of Glasgow, Glasgow, UK, 2016
Correlation Network Approaches in Metabolomic Epidemiology, Imperial College London, London, 2016
Data Analysis in Metabolomics, ECETOC - The European Centre for Ecotoxicology and Toxicity of Chemicals, Brussels, Belgium, 2016
Power Analysis and Sample Size Determination in Metabolic Phenotyping, Metabolomics 2016, Dublin, Ireland, 2016
A Workflow for Integrated Processing of Multi-Cohort Untargeted 1H NMR Metabolomics Data in Large Scale Metabolic Epidemiology, London Metabolomics Network, University of East London, London, 2016
Bayesian Deconvolution of Complex NMR Metabolomic NMR Spectra With Batman, Danish NMR Meeting, Aarhus, Denmark, 2016
Processing 1-d NMR Metabolomics Data with BATMAN, Norwegian University of Science and Technology, Trondheim, Norway, 2015
A Workflow for Integrated Processing of Multi-Cohort, Untargeted Metabolomics Data in Large Scale Metabolic Epidemiology, Metabolomics 2015, San Francisco, USA, 2015
Bayesian Modelling NMR Metabolic Profiles with BATMAN, Leibnitz Institute for Plant Biochemistry, Halle, Germany, 2015
Bayesian Deconvolution and Quantitation of 1D NMR Metabolic Profiles with BATMAN, University of Florida, Gainsville, FL, 2014
Processing 1D Metabolic NMR Spectra with BATMAN, Metabolomics 2013, Gasgow, UK, 2013
Bayesian Deconvolution and quantitation of 1-D NMR Metabolic Profiles with BATMAN, BioNMR Annual User Group Meeting, Budapest, Hungary, 2013
Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells, Imperial College London, EPSRC Workshop on Genomic Data Integration, 2013
BATMAN - Bayesian Modelling of NMR Metabolic Profiles, Workshop on Metabolomics and Proteomics, CIC bioGUNE, Bilbao, Spain, 2012
Differential Correlation Networks as Complex Phenotypes in Metabolomics, Francis Crick Institute Workshop on Systems and Synthetic Biology, Imperial College London, 2012
Differential Correlation Networks as Complex Phenotype in Metabolomics, Metabolomics 2012, 8th Annual International Conference of the Metabolomics Society, Washington DC, USA, 2012
BATMAN - A Bayesian Tool to Model Metabolomic 1-d NMR Spectra, Workshop on Data Standards in Metabolomics, European Bioinformatics Institute, Cambridge, UK, 2012
Metabolome Wide Association Studies, Metabolomics for biomedical research: where do we stand?, INSERM 214, Bordeaux, France, 2012
Mathematical Tools for Integrating Metabolomic with Other Data, Metabolomics for biomedical research: where do we stand?, INSERM 214, Bordeaux, France, 2012
Processing and Modelling of Metabolomics Data, 3rd Meeting of Danish Metabolomics SocietyUniversity of Copenhagen, Copenhagen, 2011
Bayesian Modelling of NMR Spectra for Metabolomics, Metabomeeting 2011, Helsinki, Finland, 2011
Consensus-phenotype integration of metabolomic and transcriptomic data, 3rd Sino-British Frontier Workshop of Cancer Biology / MRC-China Workshop on Cell Death, Stem Cell & Cancer, Shanghai, China, 2011
Modelling time series in metabolomics using uni- and multivariate approaches, Chemometrics' Epistemology in Systems Biology -Using the 'omics technologies to study complex biological systems, Skanör-Falsterbo, Sweden, 2011
New methods for exploring ecotoxicogenomics data: nonlinear relationships and pathway tools, An international workshop on molecular network inference: Towards a Systems Biology approach to Ecotoxicology, University of Birmingham, 2011
Chemometrics in Metabolic Profiling: Statistically Rigorous Modelling of LC-MS data, Chemometrics in Analytical Chemistry, Antwerp, Belgium, 2010
Integrating transcriptomic and metabolomic data to enhance the detection of pathways associated with drug response, Syngenta, Jealotts Hill, UK, 2010
Consensus-phenotype integration of metabolomic and transcriptomic data, American Chemical Society Metabolomics Workshop, San Francisco, CA, USA, 2010
Statistical and Bioinformatic Methods for Metabolomics, University of Reading Systems Biology Network, Reading, UK, 2010
Statistical and Bioinformatic Methods for Metabolomics, Imperial College Department of Statistics, London, UK, 2009
Statistical and Bioinformatic Methods for Metabonolomics, University College, London, UK, 2009
Statistical Methods in Metabolic Profiling, TOP Institute, Rotterdam, The Netherlands, 2009
Bioinformatic approaches for predicting toxivity and biomarker data mining in metabolic profiling, Visiongain Biomarkers Conference, London, UK, 2009
Chemometric methods in metabolic profiling, Univeristy of Mons, Mons, Belgium, 2009
Bioinformatic Methods in Metabolic Profiling Studies of Toxicity, NERC International Collaboration Inititive, University of Birmingham, Birmingham, UK, 2008
EBI Metabolomics Workshop: Imperial College viewpoint, European Bioinformatics Institute, Hinxton, Cambridge, UK, 2007
Semi-Automated Systems for Organ Specific Toxicity Screening in Metabonomics: the COMET Approach, DIA, Amsterdam, Netherlands, 2006
Bioinformatics for Metabonomics, International Conference on Magnetic Resonance in Biological Systems, Goettingen, Germany, 2006
springScape: Visualisation of Microarray and Contextual Bioinformatic Data using Spring Embedding and an ‘Information Landscape’, Intelligent Systems in Molecular Biology 2006, Fortaleza, Brazil, 2006
Metabonomic Toxicology: Expert Systems and Statistical Approaches, SciPharm, Edinburgh, UK, 2006
Bioinformatic & Statistical Approaches to Modelling of Metabonomic Data, The Preclinical Interface, London, UK, 2006
Visualisationof Genomic and Metabonomic Data on Biological Networks using Spring Embedding, Metabomeeting 2, Cambridge, UK, 2006
Chemometric and Bioinformatic Approaches to the Analysis of Metabonomic Data, HFL, Newmarket, UK, 2005
Spring Embedding for Visualisation of Post-Genomic Data on Biological Networks, Imperial College London, London, UK, 2005
The Standard Metabolic Reporting Structures Group (SMRS), Cambridge, UK, 2005
Chemometric methods for metabonomics in toxicology: Constructing an expert system for the COMET project, Scandinavian Symposium on Chemometrics 2003, Aland, Finland, 2003
Research Staff
Research Student Supervision
Cooper,A, Bayesian modelling of brominated flame retardants
Zukovski,E, Unsupervised modelling of metabolomics data

