Our primary goal is to use molecular approaches to study the regulation of intracellular iron homeostasis in the African trypanosome, Trypanosoma brucei
Why study African trypanosomiasis?
African trypanosomiasis or "African sleeping sickness" remains one of the world's most neglected diseases with ~60 million people at risk and ~5,000 new cases reported annually in sub-Saharan Africa. Available treatments are highly toxic & no vaccines currently exist. There is, therefore, an urgent need for the identification of novel therapeutic targets.
Unfortunately, there is no economic incentive from big Pharma to invest in the development of new drugs. Much hope relies on basic biomedical science research to understand the complex pattern of these infections.
The causative agent is T. brucei - a single-celled flagellated protozoan parasite that lives and multiplies in blood and lymphatic tissues of infected mammals: humans, cattle, horses, wildebest, etc. They are transmitted by insect vectors known as tse-tse flies of Glossina spp. but some species can also be transmitted mechanically. We can culture parasites both in vivo and in vitro in the lab. The availability of molecular and experimental tools: targeted gene disruption, epitope-tagging, constitutive & conditional expression, dominant-negative phenotypes, conditional RNA silencing, and “omics” approaches make T. brucei a powerful "model" system to study both evolutionarily "divergent" and evolutionarily "conserved" biological processes.
Intracellular iron regulation

Trypanosomes live extracellularly in blood where they interact with their host obtaining essential micronutrients such as iron for survival.
Iron is a critical co-factor for numerous important reactions in both eukaryotes and prokaryotes, but in the presence of oxygen iron can be very toxic - creating the need for very tight regulation of free iron levels.
In mammalian cells this regulation is achieved by iron regulatory proteins (IRP e.g. aconitase) containing iron-responsive elements (IRE) that control expression of major components of the iron acquisition pathway such as the transferrin receptor (TfR) - see (A).
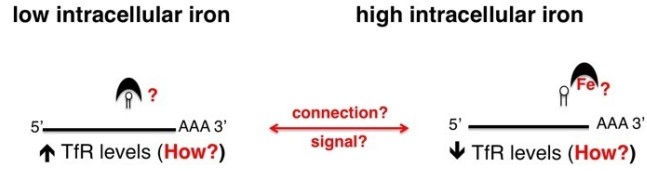
However, how this task is achieved in African trypanosomes is completely unknown (B). Interestingly, African trypanosomes have a surface protein that functions as a transferrin receptor (TfR), and expression levels of this protein is also modulated by iron availability. However, TbTfR is structurally distinct from the mammalian TfR, and previous work indicates that T. brucei lacks the canonical IRP/IRE iron-based regulatory system.
Our research will seek to understand if trypanosomes have co-opted alternative eukaryotic mechanisms for iron-sensing and regulation of iron homeostasis under three broad themes.
Ongoing Projects

Project I: Post-transcriptional regulators
Virtually nothing is known about the post-transcriptional regulatory processes that maintain intracellular iron homeostasis. We will perform genetic screens coupled with next generation sequencing to discover these new trypanosome-specific iron-responsive factors. We are currently tackling a few factors in this pathway.

Project II: Post-translational regulators
We hypothesize that novel iron-signalling effectors exist at the post-translational level that control iron uptake. We will use a proteomics approach to discover new iron-responsive sensors and signal effectors in trypanosomes. We have identified a phosphatase that may act as the iron-sensor and are currently studying its function.

Project III: Mapping the iron-regulatory network
This project systematically integrates projects I and II. Here we are performing functional studies to provide mechanistic insights into the specifics of protein-RNA, protein-protein and protein-DNA complex interactions that may regulate intracellular iron levels.
Contact us
Dr. Calvin Tiengwe
Dept. of Life Sciences,
503 Sir Ernst Chain Building,
Imperial College London,
SW7 2AZ, UK
