Overview
About me
I am a UKRI Innovation Fellow at Health Data Research UK and Clinical Reader in Rheumatology. My research combines my clinical expertise in inflammatory diseases (Consultant Rheumatologist) with data science and computational biology. I moved to Imperial College as a Group Leader in 2019.
Background and previous research

I graduated in Medicine from the University of Edinburgh in 2004. I undertook specialist clinical training in Rheumatology in London, where I developed a particular interest in lupus and vasculitis, and vascular inflammation more broadly.
I have a strong quantitative background: I have an MPhil in Computational Biology and PhD in genomics from the University of Cambridge.
i) Doctoral research
I did my PhD with Prof. Ken Smith (Department of Medicine) and Prof. Sylvia Richardson (MRC Biostatistics Unit). My doctoral research focussed on identifying genetic determinants of the transcriptome (expression quantitative trait loci, 'eQTLs') in peripheral blood immune cell subsets from patients with inflammatory bowel disease, ANCA-associated vasculitis and healthy controls. This work revealed how the genetic control of gene expression varies between cell types and between health and inflammatory disease states, and provides insight into how non-coding variants may influence susceptibility to immune-mediated diseases (Peters et al, PLoS Genetics 2016).
ii) Post-doctoral research
I did my post-doctoral research fellowship with Prof John Danesh in the Cardiovascular Epidemiology Unit at the University of Cambridge. This research involved applying high-throughput proteomic assays at epidemiological scale, integrating these data with genomic data to identify protein quantitative trait loci (pQTLs), and using this information to perform Mendelian randomisation analysis.
Protein QTL mapping
We performed pQTL mapping in 3,300 blood donors using a novel aptamer-based proteomic assay (Sun et al Nature 2018). This work identified 1,927 genetic associations with 1,478 proteins, a fourfold increase on existing knowledge. These data provide important fundamental and translational insights.
Example: identification of pQTLs for plasma proteinase-3 (PR3). PR3 is a serine protease and is the major autoantigen in ANCA-associated vasculitis (particularly the subset called granulomatosis with polyangiitis, formerly known as Wegener’s granulomatosis). We identified two independent genetic regions that influence plasma PR3 abundance in healthy blood donors: i) the region upstream of the PRTN3 gene (encoding PR3) and ii) a missense variant in SERPINA1 (encoding alpha-1 antitrypsin, A1AT, an anti-protease). We showed that genetic variation in the former influences total PR3 levels, whereas the latter results in increased free PR3 (due to less PR3-A1AT in complex). These data explain the known genetic associations with anti-PR3 vasculitis. Genetic variation that increases the autoantigen PR3 in plasma (either total PR3 of free PR3) confers increased risk of vasculitis through 2 independent mechanisms. This provides strong human evidence that puts PR3 at the centre of pathogenesis, rather than reflecting an epiphenomenon.

EGPA genetics
Churg-Strauss syndrome (aka eosinophilic granulomatosis with polyangiitis, EGPA) is a rare disease is characterised by asthma, eosinophilia, granulomatous inflammation and vasculitis. Although currently classified as a form of ANCA-associated vasculitis, only one third of patients have auto-antibodies (ANCA), almost always directed against myeloperoxidase (MPO). As part of the European Vasculitis Genetics Consortium, we conducted a genome-wide association study (GWAS) of EGPA (Lyons et al 2019). This revealed 8 genetic loci associated with the condition, with the strongest association at HLA-DQ. We showed that EGPA shares genetic architecture with eosinophil count in the general population and also asthma. I showed through Mendelian randomisation that a tendency to higher eosinophil count is a causal risk factor for the condition, rather than eosinophilia being a consequence of disease or an epiphenomenon. Through analysis stratified by antibody status, we revealed that EGPA comprises two clinically and genetically distinct syndromes. I showed that patients with anti-MPO antibodies had a higher frequency of glomerulonephritis and neuropathy, whereas ANCA negative patients had a higher prevalence of cardiac involvement and lung infiltrates. The association at HLA-DQ was restricted to the anti-MPO positive subgroup. These data show that anti-MPO positive EGPA is an autoimmune disease with a class II HLA association, whereas antibody negative EGPA may have more in common with idiopathic hypereosinophilic syndromes. This study adds to an emerging body of evidence that supports redefining ANCA-associated vasculitis by autoantibody status.
CURRENT RESEARCH PROJECTS

Current research projects
1) Protein QTL mapping and Mendelian randomisation.
I am a Principal Investigator in the SCALLOP plasma proteomics consortium, leading a pQTL discovery project project for inflammation-related proteins in ~15,000 individuals (with Adam Butterworth, University of Cambridge). As well as providing important insights into the genetic architecture of plasma proteins, pQTL data have two valuable applications for understanding human disease:
1) Identifying the effector molecules that underlie genotype-disease associations identified by GWAS. Since most variants associated with common disease lie in non-coding regions, the mechanism by which they influence disease risk is often unclear.
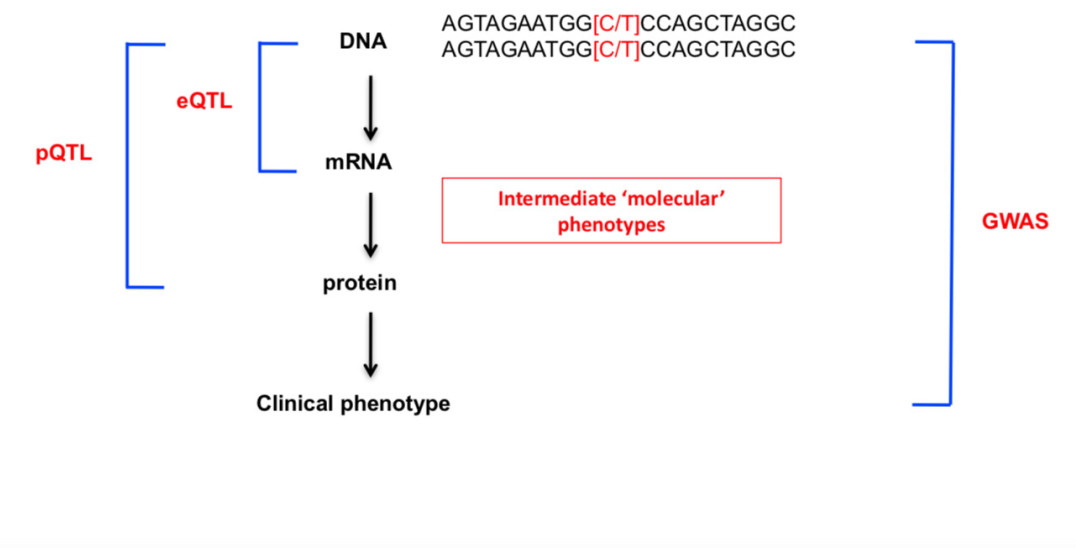
Connecting the genome to phenome through QTL mapping
2) As 'genetic instruments' in Mendelian randomisation analyses to evaluate whether particular proteins play a causal role in a range of diseases. The random allocation of alleles at meiosis effectively provides a naturally randomised trial, allowing causal inference from observational data. For example, we previously showed that genetic predisposition to higher MMP-12 is associated with lower risk of coronary disease and stroke (Sun et al, Nature 2018).
We are particularly interested in creating “genetic “dose-response” curves, that allow us to anticipate the effect of therapeutic manipulation of a protein. Such information allows putative drug targets to be prioritised/deprioritised before embarking on costing clinical trials.

This talk gives an overview of our work:
2) COVID-19 multi-omics (transcriptomics and proteomics).
We are currently performing plasma proteomics and single cell and bulk RNA-seq on peripheral blood samples from patients with COVID-19, focusing on end-stage kidney disease (ESKD) patients. ESKD patients are at high risk of severe and fatal COVID-19. These data should reveal the molecular mediators that cause a subset of individuals to develop severe disease and identify therapeutic targets and opportunities for drug repurposing.
Read our COVID-19 studies here:
Plasma proteomics in COVID-19 using Olink immunoassays:
https://doi.org/10.7554/eLife.64827
Multi-omics in COVID-19 (immune cell RNA-seq and Somascan proteomics):
3) SLE genomics and transcriptomics.
Collaborations with Prof Tim Vyse (Kings College London), Emma Davenport (Wellcome Sanger Institute) and Mike Inouye (University of Cambridge/ Melbourne Baker Institute). We are interested in understanding the influences of genetics on SLE risk and how these genetic risk variants impact on intermediate molecular traits.
4) Giant cell arteritis (GCA) and Takayasu arteritis
-Genetics, transcriptomics and proteomics (Collaboration with Prof Ann Morgan, University of Leeds).
-Novel PET-MRI imaging in large vessel vasculitis (Collaboration with Jason Tarkin and Prof Justin Mason). We are investigating whether novel PET ligands targeting macrophages provide more sensitive and specific imaging of large vessel vasculitis.
CLINICAL

I am currently an Honorary Consultant Rheumatologist at Imperial College Healthcare NHS Trust. I previously held Consultant positions at Addenbrookes Hospital in Cambridge (Lupus and Vasculitis Service) and at Barts Health in London.
Please note that the academic College (imperial.ac.uk) email is not approved or suitable for communications about patients, and such emails will not be responded to. If you are a healthcare professional or a patient requiring clinical advice, you should contact the Rheumatology Department at Imperial College Healthcare NHS Trust on 0203 311 6622.

