Summary
Dr. Veselkov’s research group focuses on developing computational/AI methods that can make a difference in data-driven global health and disease. He has led the development of a series of field-changing data analytics frameworks to augment emerging molecular ("-omics") profiling technologies into clinical decision support, and population health management. His group expertise combines the use of a diverse range of computational techniques from signal processing, imaging informatics, network-based machine learning, natural language processing and high-performance mobile computing for information extraction from heterogeneous biomedical datasets. In collaboration with Vodafone Foundation, Dr Veselkov is currently leading and managing DreamLab-DRUGS and CORONA-AI projects - one of the biggest citizen science projects with over 250K engaged users. The projects harness the idle processing power of tens of thousands of smartphones, network-based AI and large -omics data to hunt for drug-food combinations against cancer genomes and emerging COVID-19 disease. His translation interests are omics-driven precision medicine/nutrition, AI drug repositioning, and digital pathology. Dr Veselkov has received a World Economic Forum (WEF) Young Scientist Award, and served on the WEF Global Agenda Council on the Future of Computing. His research was featured by technology programs BBC Click, Sky Swipe, and numerous media outlets.
DReaMLAB- HyperFoods: Machine intelligent mapping of DISEASE-beating molecules in FOODS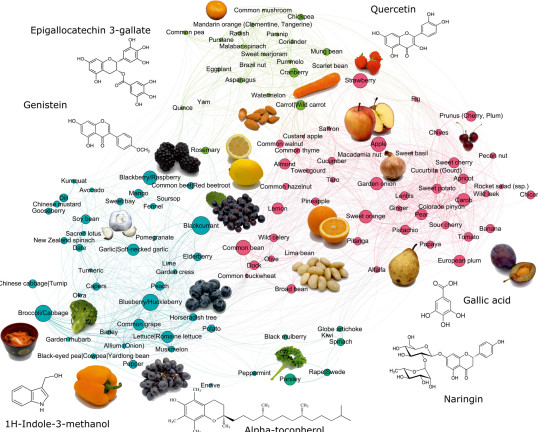
Laponogov I and Gonzales G, et al Veselkov K (2021) Network machine learning maps phytochemically rich “Hyperfoods” to fight COVID-19. Human Genomics 15, 1.
Veselkov K*, et al, HyperFoods: Machine intelligent mapping of cancer-beating molecules in foods (2019), Scientific Reports, 9, 9237. Journal Top 25 Collection.
MSHub and ChemDISTILLER: processing, sharing, annotation, comparison, and molecular networking of global mass spectrometry data.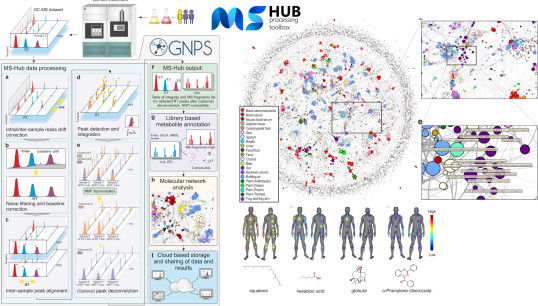
Aksenov AA, Laponogov I, et al Veselkov K* (2020) Auto-deconvolution and molecular networking of gas chromatography–mass spectrometry data. Nature Biotechnology.
Laponogov I, Sadawi N, Dieter G, Mirnezami R, Veselkov K* (2018) ChemDistiller: an engine for metabolite annotation in mass spectrometry, Bioinformatics, 34(12): 2096-2102.
BASIS: BIOINFORMATICS PLATFORM FOR PROCESSING LARGE-SCALE MASS SPECTROMETRY IMAGING IN CHEMICALLY AUGMENTED HISTOLOGY
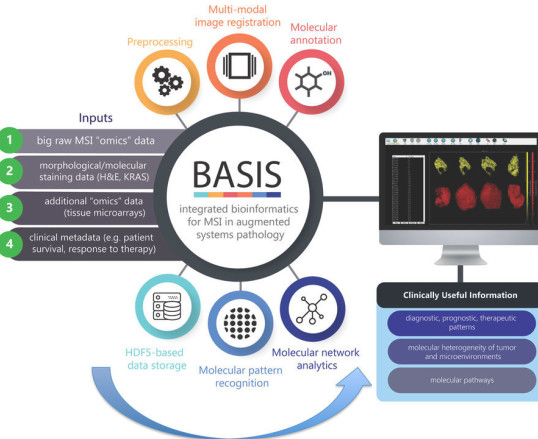
Veselkov KA*, et al (2018) BASIS: High-performance bioinformatics platform for processing of large-scale mass spectrometry imaging data in chemically augmented histology, Sci Rep, 8, 4053.
Veselkov KA*,et al (2014). Chemo-informatic strategy for imaging mass spectrometry-based hyperspectral profiling of lipid signatures in colorectal cancer, PNAS, 111: 1216-122.
HASKEE: HYPOTHESIS AND ASSOCIATIONS FROM KNOWLEDGE AND EVIDENCE NATURAL LANGUAGE PROCESSING PLATFORM
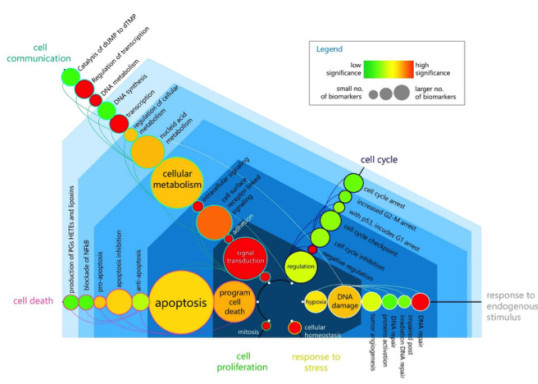 Galea D, Laponogov I, Veselkov K* (2018), Exploiting and assessing multi-source textual data for supervised biomedical named entity recognition, Bioinformatics, 34(14):2474-2482.
Galea D, Laponogov I, Veselkov K* (2018), Exploiting and assessing multi-source textual data for supervised biomedical named entity recognition, Bioinformatics, 34(14):2474-2482.
Selected Publications
Journal Articles
Gonzalez G, Gong S, Laponogov I, et al., 2021, Predicting anticancer hyperfoods with graph convolutional networks, Human Genomics, Vol:15, ISSN:1479-7364
Laponogov I, Gonzalez G, Shepherd M, et al., 2021, Network machine learning maps phytochemically rich "Hyperfoods" to fight COVID-19, Human Genomics, Vol:15, ISSN:1479-7364, Pages:1-1
Aksenov AA, Laponogov I, Zhang Z, et al., 2020, Auto-deconvolution and molecular networking of gas chromatography-mass spectrometry data, Nature Biotechnology, Vol:39, ISSN:1087-0156, Pages:169-173
Veselkov K, Gonzalez Pigorini G, Aljifri S, et al., 2019, HyperFoods: Machine intelligent mapping of cancer-beating molecules in foods, Scientific Reports, Vol:9, ISSN:2045-2322
Gu Q, Veselkov K, 2018, Bi-clustering of metabolic data using matrix factorization tools, Methods, Vol:151, ISSN:1046-2023, Pages:12-20
Galea D, Laponogov I, Veselkov K, 2018, Exploiting and assessing multi-source data for supervised biomedical named entity recognition, Bioinformatics, Vol:34, ISSN:1367-4803, Pages:2472-2482
Laponogov I, Sadawi N, Galea D, et al., 2018, ChemDistiller: an engine for metabolite annotation in mass spectrometry, Bioinformatics, Vol:34, ISSN:1367-4803, Pages:2096-2102
Veselkov KA, Sleeman J, Claude E, et al., 2018, BASIS: High-performance bioinformatics platform for processing of large-scale mass spectrometry imaging data in chemically augmented histology, Scientific Reports, Vol:8, ISSN:2045-2322
Galea D, Inglese P, Cammack L, et al., 2017, Translational utility of a hierarchical classification strategy in biomolecular data analytics., Scientific Reports, Vol:7, ISSN:2045-2322
Veselkov KA, Mirnezami R, Strittmatter N, et al., 2014, Chemo-informatic strategy for imaging mass spectrometry-based hyperspectral profiling of lipid signatures in colorectal cancer, Proceedings of the National Academy of Sciences of the United States of America, Vol:111, ISSN:0027-8424, Pages:1216-1221
Veselkov KA, Vingara LK, Masson P, et al., 2011, Optimized Preprocessing of Ultra-Performance Liquid Chromatography/Mass Spectrometry Urinary Metabolic Profiles for Improved Information Recovery, Analytical Chemistry, Vol:83, ISSN:0003-2700, Pages:5864-5872
Veselkov KA, Pahomov VI, Lindon JC, et al., 2010, A Metabolic Entropy Approach for Measurements of Systemic Metabolic Disruptions in Patho-Physiological States, Journal of Proteome Research, Vol:9, ISSN:1535-3893, Pages:3537-3544
Veselkov KA, Lindon JC, Ebbels TMD, et al., 2009, Recursive Segment-Wise Peak Alignment of Biological <SUP>1</SUP>H NMR Spectra for Improved Metabolic Biomarker Recovery, Analytical Chemistry, Vol:81, ISSN:0003-2700, Pages:56-66
Holmes E, Loo RL, Stamler J, et al., 2008, Human metabolic phenotype diversity and its association with diet and blood pressure, Nature, Vol:453, ISSN:0028-0836, Pages:396-U50



