BibTex format
@article{Serrao:2015:10.1186/s12977-015-0167-3,
author = {Serrao, E and Ballandras-Colas, A and Cherepanov, P and Maertens, GN and Engelman, AN},
doi = {10.1186/s12977-015-0167-3},
journal = {Retrovirology},
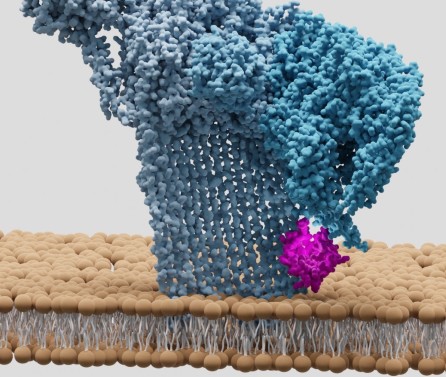
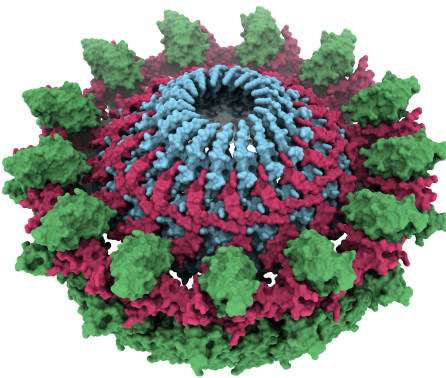
title = {Key determinants of target DNA recognition by retroviral intasomes},
url = {http://dx.doi.org/10.1186/s12977-015-0167-3},
volume = {12},
year = {2015}
}