Transition paths in state space to infer, segment and predict patient trajectories in progressive diseases
Dr Iain Johnston (Biosciences, University of Birmingham), Prof Mauricio Barahona (Mathematics) and Prof Charles Coombes (CRUK Centre & Dept of Surgery and Cancer)
Others: Dr Nicholas Kantas (Mathematics) and Prof Ara Darzi (IGHI & Dept of Surgery and Cancer),
In many progressive diseases, decisions along the patient journey are limited by the lack of tools that take into account information about the route the disease will take in individuals. This results in a blanket application of drug treatments and monitoring schemes, such that many patients receive therapies that have no effect, and others who could potentially benefit receive inadequate treatment. Similar issues occur with patient cohorts assigned to follow-up programmes in NHS organisations where longitudinal administrative data could allow the analysis of temporal trends for better prediction of care events such as emergency admissions, clinical complications or deterioration in frail elderly, to create more targeted and timely interventions based on multiple discrete categories taking into account the importance of co-associations.
This project will aim at developing tools to aid towards a richer segmentation of the population, not based on individual characteristics but instead by likely trajectories in a disease trait space. The programme will develop methods for the inference of dynamic paths in lattice-like transition networks capturing different discrete disease states. This framework will build prescriptive transition graphs describing the probabilistic progression of disease, including memory and non-Markovian dependencies. This framework, coupling a hidden Markov model approach with Bayesian inference, will be used to find and classify groups of patients exhibiting similar trajectories through the phenotypic landscape of disease. This work will be linked to state-space sampling representations of dynamical data and to community detection algorithms based on Markov paths on graphs as a means to detect groups of patients with similar features, and hence to propose specific treatment strategies based on individual history and predicted future progression.
This work will be done in close collaboration with Ara Darzi’s group at the IGHI and the CRUK Centre (Coombes) and will be linked to data curated and collected by these and other groups connected to clinical trials and NHS-wide datasets.
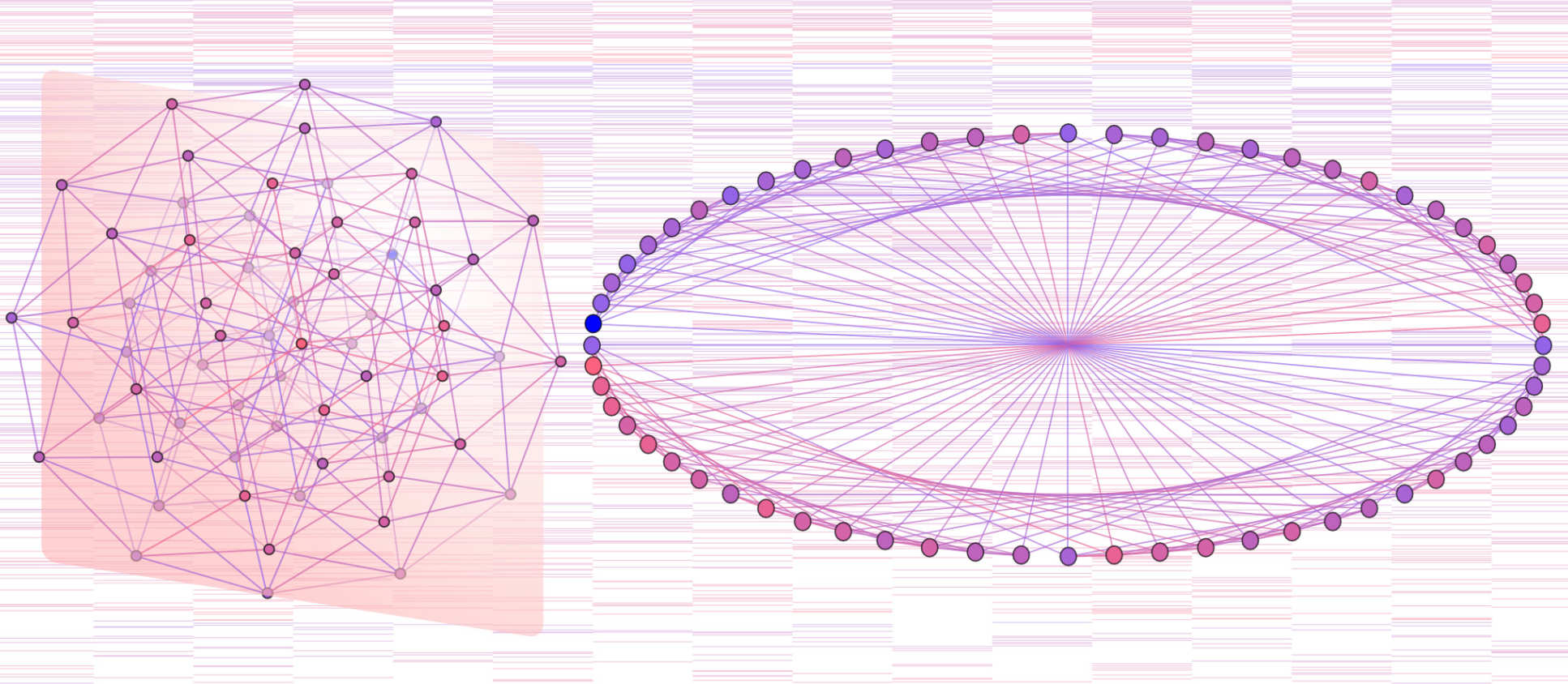
Hypercube transition networks model all possible pathways for disease progression. Patient data can characterise these networks, discovering different disease dynamics, critical regions of state space, and predicting future progression.
The Team
Team leaders
Dr Iain Johnston
/prod01/channel_3/media/migration/research-groups/johnston-iain--tojpeg_1469782411189_x4.jpg)
Dr Iain Johnston
Mathematics
Prof Mauricio Barahona
/prod01/channel_3/media/migration/research-groups/icimages--tojpeg_1469625694734_x4.jpg)
Prof Mauricio Barahona
Mathematics
Prof Charles Coombes
/prod01/channel_3/media/migration/research-groups/coombes--tojpeg_1469625805484_x4.jpg)
Prof Charles Coombes
Medicine
Others
Dr Nicholas Kantas
/prod01/channel_3/media/migration/research-groups/kantas--tojpeg_1469626097139_x4.jpg)
Dr Nicholas Kantas
Mathematics
Prof Ara Darzi
/prod01/channel_3/media/migration/research-groups/darzi--tojpeg_1469626194194_x4.jpg)
Prof Ara Darzi
Medicine
--tojpeg_1538046453332_x4.jpg)
