Our group investigate changes in DNA associated with ageing
The aging clock can be reversed, restoring characteristics of youthfulness to aged cells and tissues. Our research aims at understanding how DNA packaging and structure change as we age and whether we can target ageing related structural features to prevent agein related disorders such as Dementia and Cancer.
In the Di Antonio group we exploit a range of chemical biology tools alonside genomics to investigate the fundamental role of DNA secondary structures and epigenetic modification in ageing biology. Our mission is to identify novel targets to exploit for therapeutic intervention of rare accelerating ageing syndromes, such as Cocakyne Syndrome, and promote healthy ageing across the life course.
We are based at the Department of Chemistry and at the Francis Crick Insitute.
Research themes
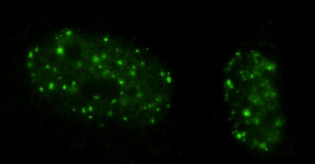
Our research on Cockayne Syndrome focuses on investigating the relationship between G-quadruplex (G4) DNA structures and this disease. Cockayne Syndrome B (CSB) is as an endogenous protein that was previously suggested to resolve G4-structures.1 We recently identified a new property of the CSB protein that might explain the premature ageing phenotype observed in Cockayne Syndrome patients. We reported a selective and astonishing affinity of CSB for a particular subtype of G4s structures, called intermolecular G4s, which can form from multiple DNA (rDNA) strands coming together in the nucleus of a cell. Although the ability of intermolecular G4s to form in vitro has been known for decades, their formation in vivo has been subject of debate due the low probability of distal DNA strand coming together to form a G4 in the context of chromatinised genomic DNA2. Our lab has reported CSB as the the first endogenous protein able to selectively recognise intermolecular G4s over intramolecular G4s (see Figure below), suggesting that these structures could indeed fold in the nucleoli of human living cells, which we further supported using Immuno-Fluorescence.3
Based on this we propose a model by which CSB is preserving cellular homeostasis by selectively interacting with intermolecular G4s. We currently investigate the structural biology aspects of this interaction and aim to map in cells the intermolecular G4s that are bound by the protein.

- Scheibye-Knudsen, M., et al., Natl. Acad. Sci. USA; (2016).
- Kolesnikova, S. and Curtis, E.A., Molecules; (2019).
- Liano, D et al., J. Am. Chem. Soc; (2021).
DNA is one of the most targeted biomolecules as it is involved in the regulation of many biological processes, including cell-growth and gene-expression. Although targeting of the canonical double helical form of DNA has been extensively investigated in the past, increasing interest is growing around the targeting of non-canonical DNA structures. Among these, DNA G-quadruplex (G4) structures are of particular interest as recent evidence suggests that they could be involved in regulating gene expression. Despite the increasing number of scientific observations demonstrating the relevance of G4s in various biological processes, there is still a significant amount of skepticism about the actual relevance and existence of these structures in living cells.
Our group uses chemistry and genomics to target and identify DNA G-quadruplex in human cells but also in parasites, such as T. Brucei ( in collaboration with Dr Monti). An example on how we leverage chemical biology tools to target G4s and study their biological significace can be taken from our study on the use of LNA-modified oligonucleoties to target and disrupt G4-structures in cells ( see Chowdhury et al. Nucleic Acids Res. 2022).
Similarly, our approach on the use of genomic tools to understant the biological significance and explore the therapuetic potential of G4-structures has been reviewed in Robisons et al. Nucleic Acids Res. 2021. Generally speaking we are eager to develop novel chemical biology and genomic platform to interrogate cells about the role of DNA secondary structures, especially in the context of epigenetic regulation and ageing.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are just two of the many neurodegenerative diseases characterised by the presence of pathological aggregates. Despite the first symptoms got related to the diseases back in the second half of the 1800s century, the mechanism by which such aggregates come to exist has not yet been thoroughly c haracterised and treatment for such pathologies is mostly ineffective. Whilst most of neurodegenerative diseases are characterised by protein-based aggregates, ALS and FTD feature a significant contribution by RNA molecules. Indeed, one genetic trait that is characteristic of this desease is the expantion of the repeat (GGGGCC) in the gene C9Orf72.
haracterised and treatment for such pathologies is mostly ineffective. Whilst most of neurodegenerative diseases are characterised by protein-based aggregates, ALS and FTD feature a significant contribution by RNA molecules. Indeed, one genetic trait that is characteristic of this desease is the expantion of the repeat (GGGGCC) in the gene C9Orf72.
Recent studies have highlighted the fundamental role of nucleic acids in phase separation events in genes associated with the diseases, offering a potential alternative to protein-lead aggregation. In particular, repeat gene expansion that are associated with the disease have shown to phase separate in their own right AND to form alternative DNA secondary structures like G-quadruplexes (G4s).
Our research in this space aims to investigate the role of nucleic acids secondary structures in the process of aggregation that causes ALS and FTD. Our recent report of the first endogenous protein that selectively recognises inter-molecular G4-structures (Liano et al. J. Am. Chem. Soc. 2021) suggests that intermolecular G4s might be formed in a cellular context and might play a role in the formation of such aggregates. In collaboration with Dr Di Michele and Dr Elani at Imperial College, we investigate using standard biophysical methods, if the formation of such secondary structures can lead to the formation of pathological aggregates.