Much of our work addresses the imaging and measurement of disease mechanisms in cell cultures, in model organisms and in patients or in ex vivo tissue. We particularly aim to exploit the molecular specificity of optical readouts to study biological processes and to identify signatures of disease. When imaging or undertaking measurements in biological samples, key challenges for biophotonics include contrast, spatial resolution/precision and imaging/sampling depth.
For thin/transparent samples such as 2-D cell cultures on a microscope coverslip, the spatial resolution is limited by diffraction and the main challenges and opportunities for biophotonics are associated with contrast. Intensity-based measurements can provide information about the distribution of chromophores through their absorption or fluorescence properties and time resolved imaging can reveal spatio-temporal dynamics, e.g. of specific labelled proteins. This is particularly powerful when genetically expressed chromophores such as fluorescent proteins are employed, since they permit cell signalling mechanisms to be visualised in live cells in real time. Further information can be provided by spectroscopic analysis. For absorption or fluorescence readouts, spectral resolution can permit unmixing of different molecular components, for example, oxy- and deoxy-haemoglobin can be separated through their different absorption spectra to permit in vivo measures of blood oxygenation and multiple fluorophores can be separated through differences in their excitation and/or emission spectra or their fluorescence lifetime. Further information about the local fluorophore environment can be extracted from changes in the fluorescence emission parameters.
While microscopy of cells on glass coverslips has been enormously important for the development of our understanding of cell biology and the discovery of new drugs and will continue to play a crucial role, including in higher throughput multiwell plate systems for HCA, there is increasing concern that biological processes observed in monolayers of cultured cell are not necessarily reproduced in live organisms – or human patients. This is of crucial importance to drug discovery and testing but practical constraints – particularly the desire for convenient readouts – continue to constrain most assays to be undertaken in conditions that are far removed from their native or intended physiological contexts. There is therefore an increasing trend to translate fluorescence assays from cell monolayers to supposedly “more realistic” three dimensional cell and tissue cultures, to transparent live organisms that are amenable to genetic manipulation, such as drosophila and zebrafish, to small mammals and ultimately to patients. Optical sectioning microscopes can be used to image biological processes in 3-D cell cultures and we are developing automated multiwell plate microscopy systems for assays of 3-D cell cultures such as tumour spheroids using optical sectioning FLIM and OPM.
There is also increasing interest in applying optical sectioning techniques for 3-D imaging of biological processes in small model organisms such as C. elegans and zebrafish larvae. While confocal/multi-photon laser scanning microscopes can provide optical sectioning to permit the acquisition of 3-D images and also offer improved contrast compared to wide-field imaging, they suffer from limited (100’s µm) penetration depth due to optical scattering and usually image over relatively small fields of view (10’s – 100’s µm) such that the acquisition of 3-D data sets can be very time consuming for samples on a mm scale. Confocal and multiphoton laser scanning microscopy also exhibit anisotropic resolution (typically axial resolution is significantly worse than transverse resolution). For larger samples in the “mesoscopic” regime (~0.5-10 mm), a number of imaging techniques including OPT and light sheet microscopy techniques are being utilised to provide new insights into structure and function of whole organisms. In vivo imaging of larger samples, however, is compromised by the scattering of optical radiation and techniques to address this are being actively developed using ballistic light techniques and adaptive optics to compensate for aberrations and scattering. As intensity imaging becomes degraded due to scattering in larger samples, it is still possible to exploit quantitative readouts of biological function using spectroscopic techniques such as FLIM, which has been used map FRET in live zebrafish in combination with OPT1.
For thicker biological tissue samples images must be formed utilising ballistic light near the tissue surface or using scattered light for tomography. The former approach can be extended using OCT or multiphoton microscopy, which can be applied to animal models or to patients. We have worked on an in vivo multiphoton FLIM platform for clinical studies and have applied it to the diagnosis of skin cancer. Endoscopy can be used to couple optical radiation to tissue surfaces that are not accessible. There is also considerable interest in developing tomographic techniques for non-invasive readouts of biological function. We are developing diffuse fluorescence tomography for murine disease models and have worked with the MRC Clinical Sciences Centre to demonstrate FLIM FRET of genetically expressed fluorescent proteins in live mice and are working with UCL to develop a tomographic fluorescence imaging platform for imaging disease processes in live adult zebrafish for studies of cancer, inflammation and bacterial infection. We are also starting a programme to apply photoacoustic tomography with DFT and OPT to study epigenetic gene expression in murine models of disease.
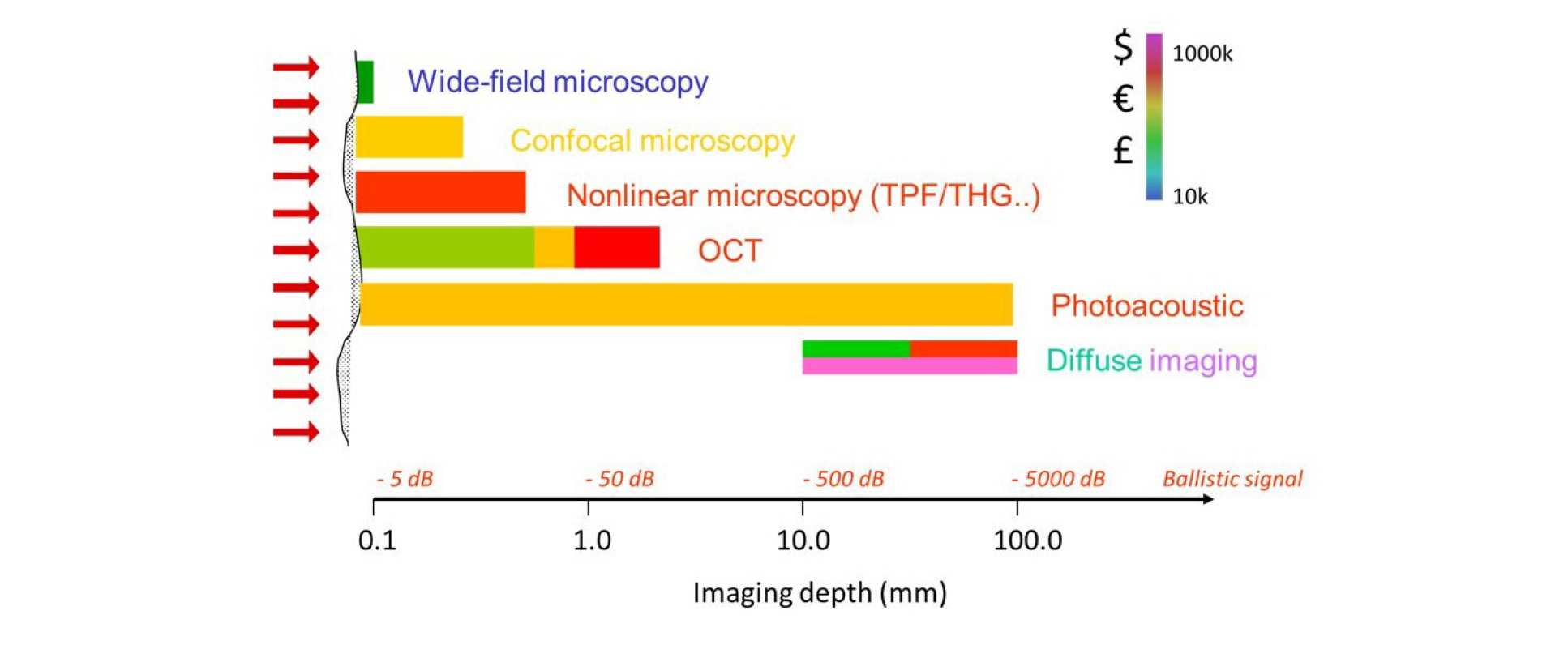
The figure below summaries the range of optical imaging techniques and their scope for application, in terms of imaging depth. The diagram is colour coded in terms of the cost of the different approaches. Some approaches, such as OPT or diffuse light imaging, can be implemented at relatively low cost but have also been employed in more complex, expensive approaches to enhance performance.
1 N. Andrews et al., Early view J. Biophotonics 1–11 (2016) / DOI 10.1002/jbio.201500258