A major component of our biophotonics research portfolio concerns the application of optical technology to the study of disease and the development of diagnostic tools and therapies. These biophotonics projects particularly utilise fluorescence readouts and span cuvette-based solution studies of molecular biology, super-resolved microscopy, high content analysis (HCA), preclinical imaging and clinical studies involving patients. This is an exciting time for biophotonics as traditional barriers to observation are being pushed back and life scientists can envisage learning about biomolecular processes with unprecedented detail, speed and physiological relevance. We aim to leverage our expertise in photonics technology to create new tools and opportunities for molecular cell biology and drug discovery. Our vision is to provide higher-content analysis at all scales of measurement and to make these tools as widely available as possible by developing low-cost instrumentation with open-source software.
For solution-based measurements of biomolecular interactions we have developed a novel automated multidimensional fluorometer that is able to analyse fluorescence signals with respect to excitation and emission wavelength, fluorescence lifetime and polarisation in a single experiment. This may be used for cuvette-based studies and it can be configured to automatically acquire data from multiwell plates or coupled to a fibre-optic probe for remote measurements, e.g. of biological tissue. This capability has been applied to label-free studies of cancer, heart disease and osteoarthritis.
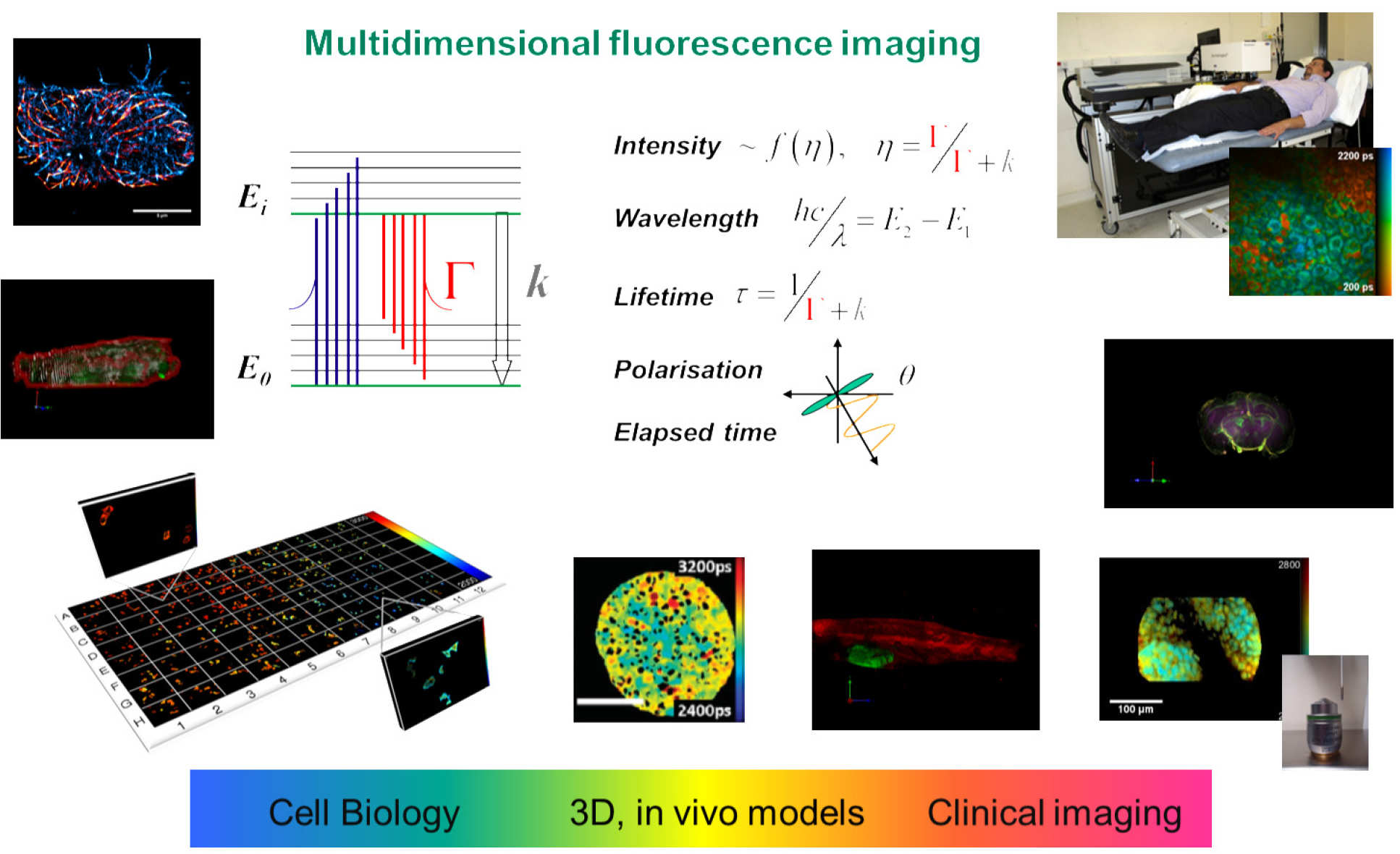
For imaging of cells and biological tissue, we have developed a wide range of fluorescence microscopes, including conventional wide-field and total internal reflection fluorescence (TIRF) microscopes – which we have extended to single molecule localisation microscopy for super-resolved imaging – and laser scanning confocal/multiphoton microscopes, which including tandem scanning (Nipkow disc) instruments for rapid live cell studies, stimulated emission depletion (STED) microscopy for super resolution and multiphoton microscopy for clinical and other applications. We have also developed light-sheet microscopy and optical projection tomography (OPT) for 3D imaging of cellular and mesoscopic samples, complemented by endoscopes and intravital microscopes for preclinical and clinical imaging.
We have a particular interest in fluorescence lifetime imaging (FLIM) and have implemented FLIM with almost all of our imaging modalities. We also resolve fluorescence signals with respect to excitation and emission spectra and polarisation, realising a multidimensional fluorescence imaging (MDFI) technology platform for biomedical research. Working with our colleagues in biology and medicine, we particularly apply our MDFI technology to measure Förster resonant energy transfer (FRET) of protein-protein interactions in order to provide information on cell signalling processes for drug discovery and fundamental studies of molecular disease mechanisms. We have developed an open source MATLAB-based tool called FLIMfit for the analysis of FLIM data that provides unprecedented functionality, including global fitting of complex decay profiles, polarisation-resolved FLIM data, time series and FLIM data from multiwell plate arrays.
For drug discovery and for basic research in the life sciences, there is an increasing trend towards higher throughout automated imaging – sometimes described as high content analysis (HCA). The automated acquisition of 100’s to 1000’s of fields of view can provide image data for statistically robust assays in a few hours, removing operator bias and accelerating discovery and screening far beyond what is possible with manual microscopy. To this end we first developed automated FLIM multiwell plate readers that require only a few minutes to each FOV, including all sample translation and focussing steps. Together with associated analysis software, this technology makes FLIM a practical tool for HCA of fixed or live cell assays. We have applied this FLIM HCA technology to both heteroFRET and homoFRET readouts of protein interactions – including of fluorescently-tagged endogenous proteins – and label-free readouts of changes in cellular metabolism utilising FLIM of NAD(P)H autofluorescence. We have also invented and developed a light-sheet microscopy technique called oblique plane microscopy (OPM) that works with established microscopes and multiwell plate readers and acquires volumetric image data at video rate (25 vol/s). This enables the imaging of dynamic events in live cells and the quantification of dynamics - including cell morphology and migration – at scale in multiwell plate screens. This OPM platform is particularly powerful for imaging 3D cell cultures and is being applied to high throughput assays of 3D melanoma cell dynamics. We are developing an open source approach to HCA that can be implemented on commercial or in-house built microscopes and most recently are applying this to high-throughput super-resolved microscopy.
Our work on super-resolved microscopy began with STED microscopy, for which we built the first super-resolved microscope to incorporate time-resolved detection for FLIM and to gate out non-super-resolved signals. This instrument also included a spatial light modulator (SLM) for the first demonstration of electronic aberration correction and PSF engineering in a STED microscope. It also demonstrated the use of a mode-locked Ti:Sapphire laser to provide the depletion laser beam and to provide tunable excitation through spectral selection of a supercontinuum generated in micro-structured optical fibre. We extended this approach to 3D STED, using the SLM to provide a 3D depletion beam and to compensate optical aberrations in the sample. Recently we further extended the approach to develop easySLM-STED – a SLM-based technique that provides easy alignment of excitation and depletion beams, significantly increased super-resolved fields of view and multiplexed excitation/depletion beam pairs for parallelised imaging. We have also worked on and stochastically-switched single molecule localisation microscopes (SMLM) based on PALM and STORM and are developing an automated high-throughput STORM microscope for super-resolved assays/screens and single particle averaging. We initially developed easySTORM – a novel approach to utilise low-cost high-power multimode diode lasers for TIRF and STORM over large fields of view – and complemented this with a novel approach to accelerate the analysis of SMLM data using ImageJ-compatible software tools implemented in parallel on an HPC cluster.
For preclinical imaging we are keen to translate our cell-based assays to ex vivo and in vivo studies. Accordingly, we are developing optical projection tomography (OPT) instrumentation for optically cleared/transparent tissue samples and for in vivo imaging of zebrafish (from larvae to adults). For in vivo imaging we have developed a compressive sensing technique that accelerates the acquisition of OPT data – which is vital for live subjects under anaesthetic – and we have accelerated the reconstruction of such data using deep learning. In zebrafish larvae, we have demonstrated 3D cell tracking using OPT for in vivo migration studies and we have implemented FLIM with OPT to realise 3D FLIM/FRET imaging of live zebrafish expressing FRET biosensors. We have also developed diffuse optical tomography with FLIM, which we have applied to readout out a FRET biosensor expressed in vivo in mice. For higher resolution preclinical functional imaging, we have also developed FLIM endomicroscopy that we have used to quantify drug-target engagement in a murine cancer model in vivo with single cell resolution using FLIM/FRET. Thus, we have developed a range of instruments that can utilise the same FLIM FRET assays in solution, in cell culture and in vivo.
For clinical applications, multiphoton microscopy, FLIM and MDFI can be applied to autofluorescence to provide label-free molecular contrast in biological tissue. We have developed multiphoton microscopes for clinical applications including the investigation of the potential of ex vivo and in vivo FLIM with a view to developing diagnostic tools. To this end we have developed wide-field FLIM endoscopes and single point fibre-optic multidimensional fluorescence probes to provide more detailed information on complex spectro-temporal autofluorescence signals and have applied these to ex vivo and in vivo patient tissue. We have also implemented multispectral FLIM for the clinical DermaInspect™ multiphoton tomography instrument, which we applied to ex vivo and in vivo studies of skin cancer and we have developed a hand-held multiphoton microscope with built-in motion compensation to address patient motion. We have also worked extensively towards an ultracompact multiphoton endomicroscope utilising a proximal SLM to achieve scanning and focussing with no distal components.
We are increasingly working towards making our capabilities openly available for other academic research groups to replicate and develop. To this end we are making use of open source software where practical and provide versions of our software tools for download and non-commercial use. Our microscopes and other instruments were originally controlled using LabVIEW, but we are increasingly using mManager and Python for instrument control and data acquisition. Our data analysis is typically undertaken using ImageJ or MATLAB. Access all our open source software tools here.