This work on SLM-STED microscopy was supported by the BBSRC, EPSRC and the MRC, including the Next Generation Optical Microscopy initiative that supported a major project with the London Institute for Medical Sciences.
Super-resolved microscopy
SLM-STED microscopy
Our work in the Photonics Group on super-resolved microscopy began with STED microscopy in 2008, when we independently developed one of the first super-resolving microscopes in the UK. To provide flexibility, this introduced the use of a femtosecond Ti:Sapphire laser to provide the depletion pulses and to provide tunable excitation pulses via supercontinuum generation in a microstructured optical fibre, followed by spectral selection. To engineer the required “doughnut” PSF profile, diffraction from computer-generated holograms on a spatial light modulator was employed for the first time in a STED microscope, which enabled electronic adjustment of the beam profiles and also enabled the correction of optical aberrations. Using this SLM-STED technique [1] we reported the first programmable correction of aberrations in a STED microscope. We also implemented time-resolved detection for the first time in a STED microscope, using TCSPC to enable us to remove the earliest detected light that was not super-resolved and also to realise fluorescence lifetime imaging. This instrument was applied to cortical actin at artificial immune synapses activated on glass coated with Fc-MICA and ICAM-1 to understand the organization of actin at various stages during the activation.
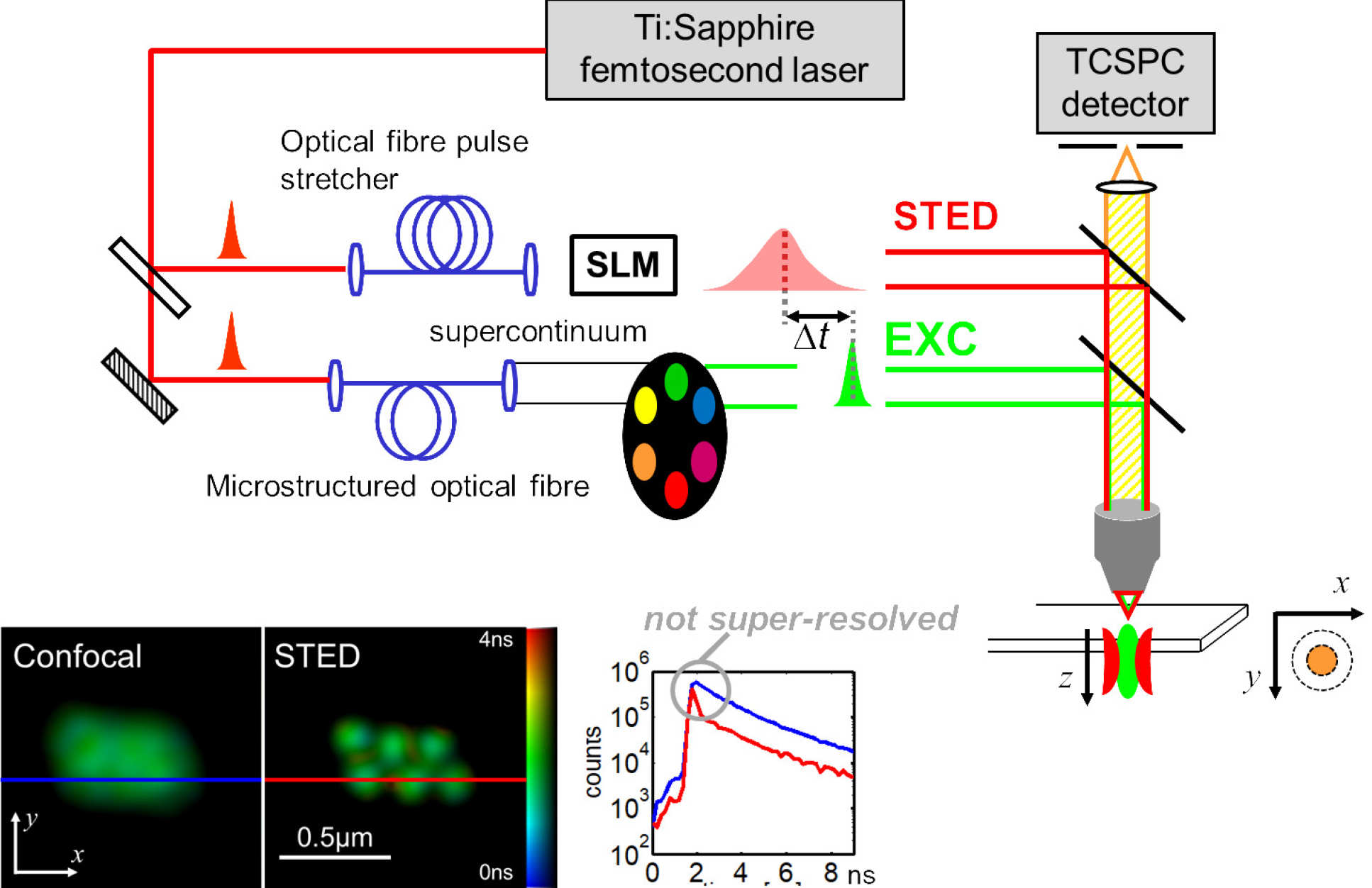
SLM-STED with Ti:Sapphire laser provided supercontinuum enabled excitation and depletion beams and time-resolved detection providing STED FLIM and gating of non-super-resolved light [1]
We subsequently extended this 2D super-resolution approach to 3D SLM-STED in 2013 in 2013 [2] using the SLM to shape the depletion beam PSF in 3D by combining the standard “doughnut PSF with a “bottle-beam” PSF. These two PSF profiles were generated by multi-passing two 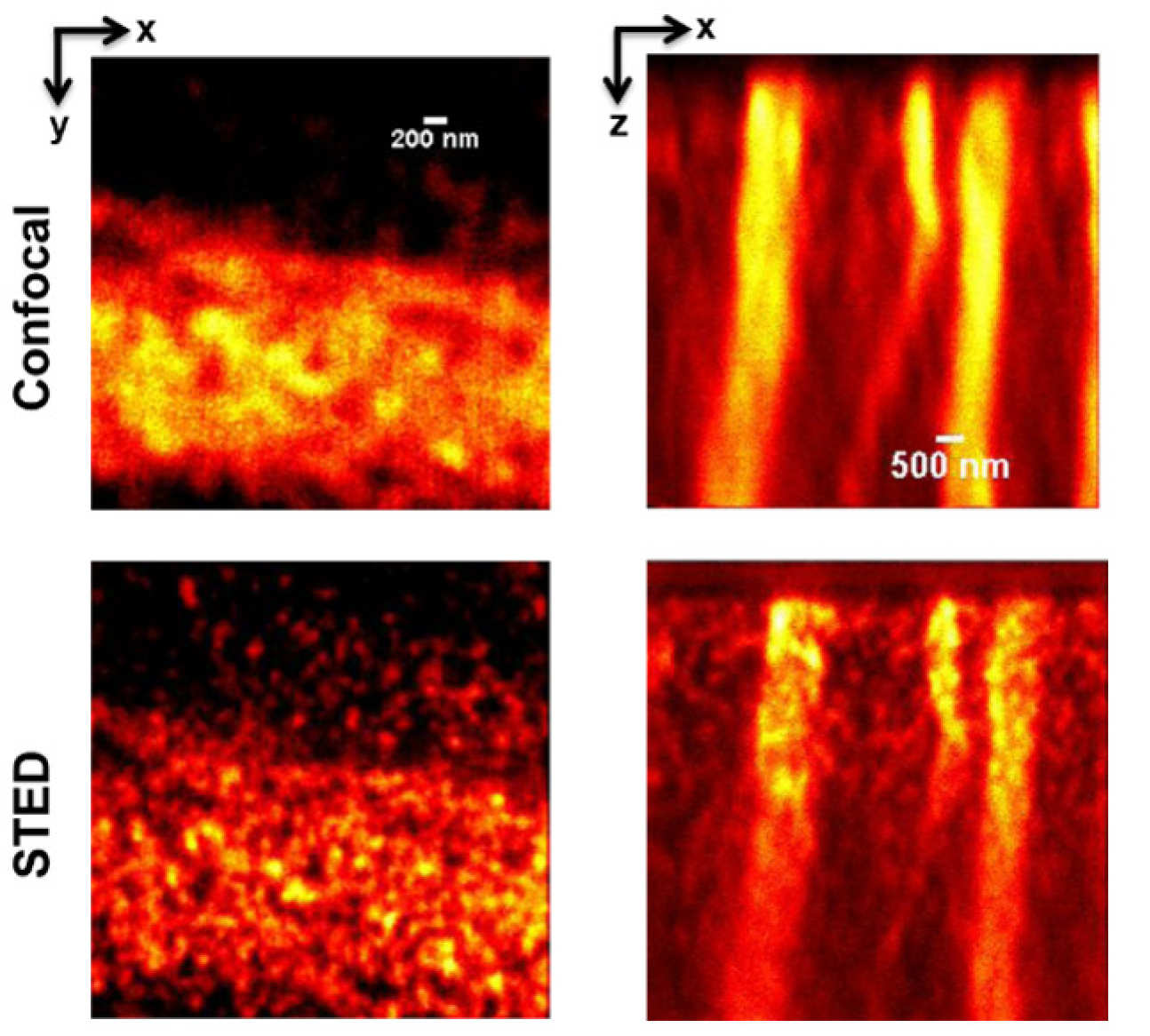 time-delayed STED depletion beams onto different regions on the SLM where different computer-generated holograms were displayed. By adjusting the power in the two beams, we could tune the depletion strength, and therefore the spatial resolution, in the lateral and axial directions. We reported this system in our second STED paper [2], where we also demonstrated the ability of the SLM to compensate for optical aberrations in the sample, which we calculated a priori and demonstrated enhanced super-resolved imaging depth in glycerol and diamond. This instrument was applied to 3D super-resolved imaging of tubulin at the immunological synapse between an NK cell and a target cell and to NV- colour centres in diamond.
time-delayed STED depletion beams onto different regions on the SLM where different computer-generated holograms were displayed. By adjusting the power in the two beams, we could tune the depletion strength, and therefore the spatial resolution, in the lateral and axial directions. We reported this system in our second STED paper [2], where we also demonstrated the ability of the SLM to compensate for optical aberrations in the sample, which we calculated a priori and demonstrated enhanced super-resolved imaging depth in glycerol and diamond. This instrument was applied to 3D super-resolved imaging of tubulin at the immunological synapse between an NK cell and a target cell and to NV- colour centres in diamond.
3D SLM-STED with adaptive optics correction of aberrations to enable 3D super-resolution of NV- centres in diamond [2]
The techniques outlined above rely on a critical alignment of the excitation and depletion beams along sperate optical paths before precisely co-aligning them in the back aperture of the objective lens. This alignment can drift over time and compromise performance. To make STED microscopes much easier to align and optimise, we developed easySLM-STED [3] which only requires users to couple both excitation and depletion laser beams into a single-mode optical fibre to maintain alignment and then a novel configuration in which the SLM shapes and directs both the excitation and depletion laser beams and delivers them co-aligned to the microscope objective lens. This configuration enables electronic adjustment of the alignment (which could be automated) and the SLM can be programmed to correct for chromatic aberration which leads to beam walk-off and limits the field of view of most STED microscopes. We demonstrated that easySLM-STED can increase the super-resolved field of view to ~100x100 mm. The flexibility conferred by using the SLM to control both the excitation and depletion beams with computer-generated holograms also provides the potential to generate multiple pairs of coaligned excitation/depletion beams for parallelised STED microscopy – providing faster super-resolved imaging. We used this approach to accelerate the STED imaging rate by a factor of 4.
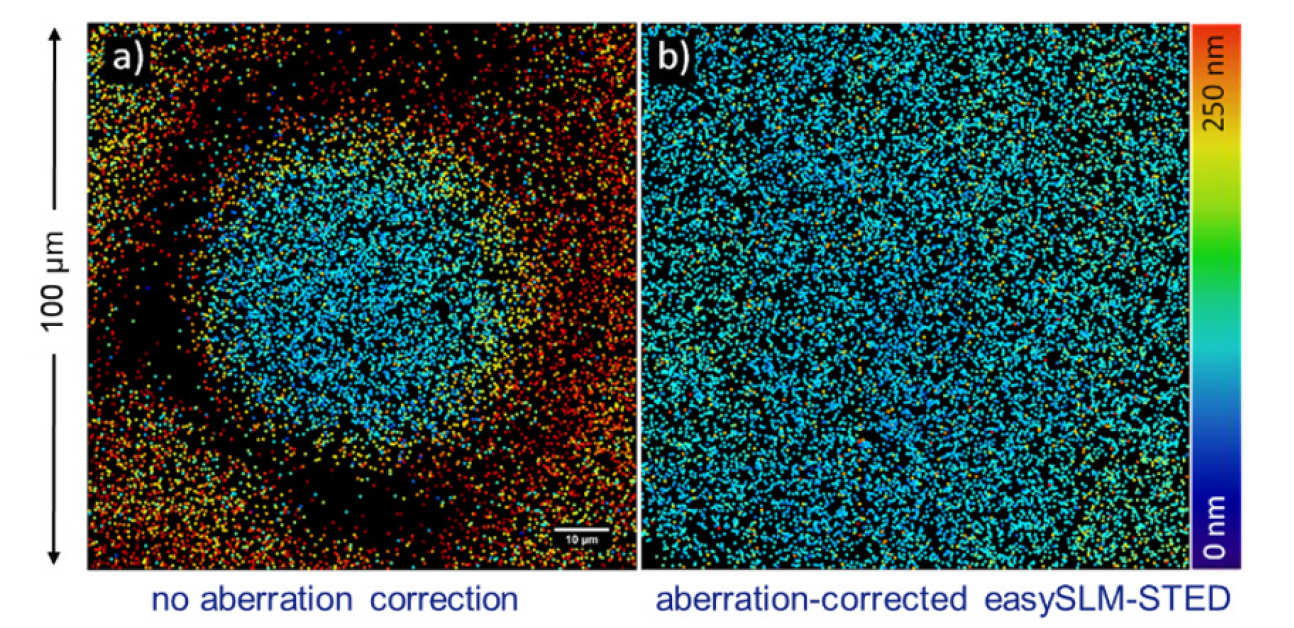 False colour scale plot of super-resolved PSF FWHM across a field of view of 40 nm fluorescent beads without and with correction of walk-off between excitation and depletion beams in easySLM-STED microscope [3].
False colour scale plot of super-resolved PSF FWHM across a field of view of 40 nm fluorescent beads without and with correction of walk-off between excitation and depletion beams in easySLM-STED microscope [3].
Structured illumination Microscopy
We have worked with a commercial SIM microscope (Carl Zeiss, Elyra S1), which is specified to provide 120 nm/300 nm lateral/axial resolution. For many studies, it is desirable to probe spatial organisation and dynamics on a scale much below 10 nm. FRET can be used to provide information of spatial scale <~10 nm, for which FLIM can provide a robust readout. FLIM/FRET can also provide functional information via FRET biosensors. We were motivated to combine FLIM with super-resolved microscopy and wanted to achieve faster imaging than we achieved with STED FLIM. Accordingly, we developed a SIM+FLIM microscope [4] that integrates SIM and optically sectioned (Nipkow disc) FLIM in a single instrument imaging the same field of view. This enables nanostructure resolved to ~120 nm to be correlated with FLIM/FRET-resolved dynamics or function. We have applied this instrument to study aggregation of the discoidin domain receptor 1 (DDR1) upon stimulation by collagen and DNA compaction during the cell cycle [4].
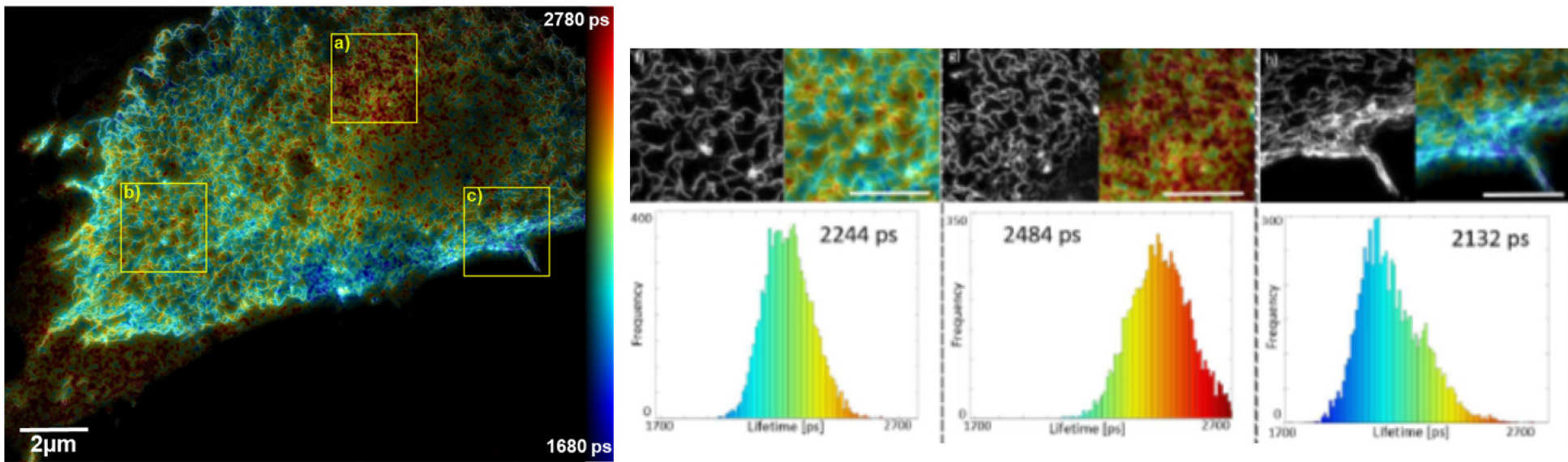
SIM+FLIM microscope configuration with SIM+FLIM maps of FRET of DDR1 receptors stochastically labelled with donor and acceptor fluorophores.
Single molecule localisation microscopy
For SMLM, we developed a robust and low-cost approach to implement total internal reflection fluorescence (TIRF) microscopy on any inverted fluorescence microscope that utilised a multimode optical fibre for efficient light coupling and the opportunity to average speckle (by vibrating fibre and mixing modes) for reasonably uniform illumination. We combined this with low-cost, high-power multimode laser diodes to provide high power (~1 W) excitation and implemented STORM using chemical buffers or using Vectashield to mount samples labelled with dyes or Citrine fluorescent protein in a technique we describe as easySTORM [5]. This can be adapted to 3D STORM using a cylindrical lens in the emission path to provide an astigmatic PSF. The figure below shows (a) TIRF and TIRF STORM images of actin cytoskeleton in fixed NKL cells with Alexa647 labelling actin mounted in STORM buffer, (b) 3-D STORM image (with colour encoded depth) of actin in HEK cells labelled with phalloidin Alexa647 and (c) a STORM image of THP-1 cells with alpha-tubulin labelled with AF647 that have been incubated with AF488-labelled S. pneumoniae.
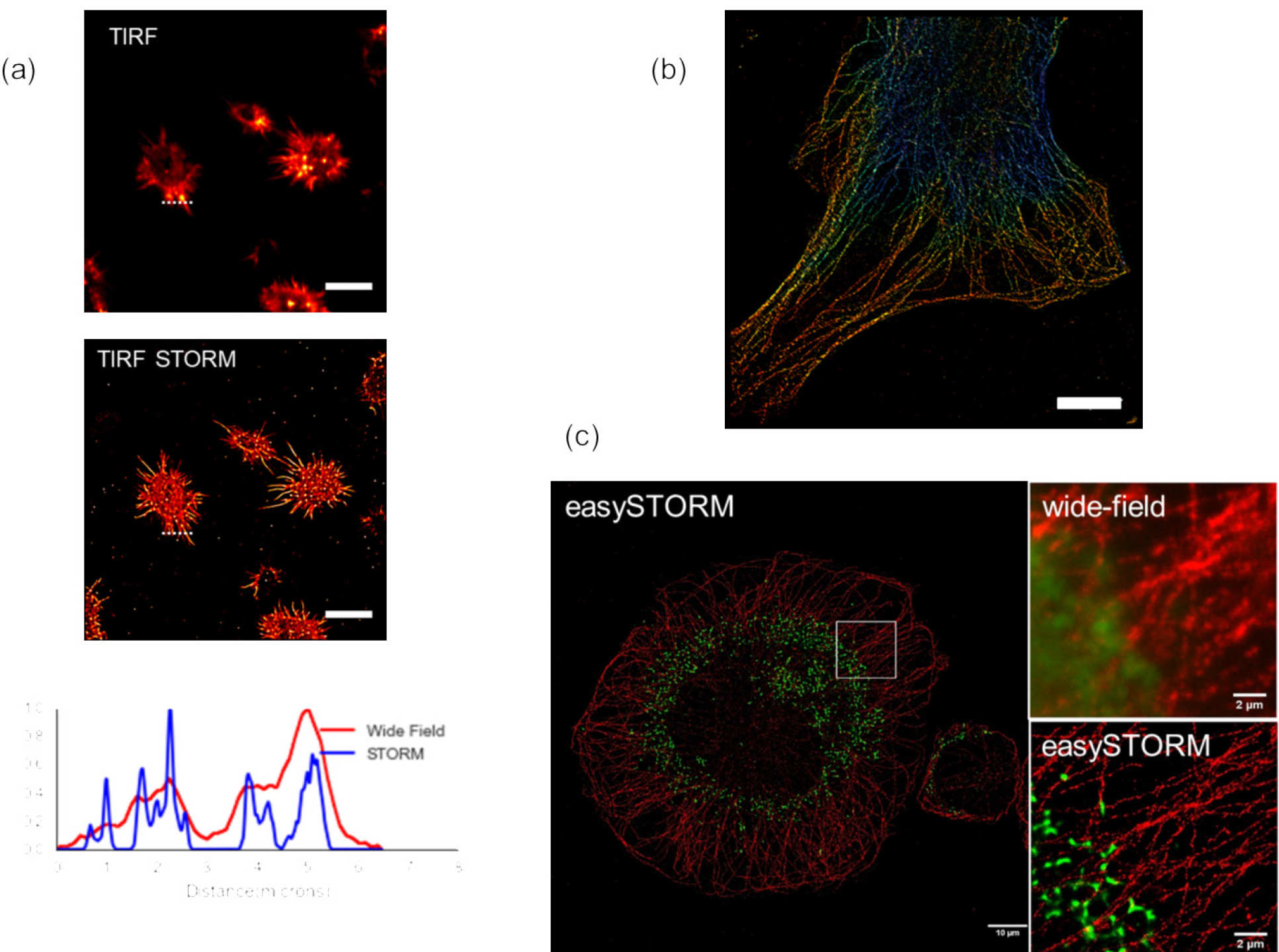
The use of high-power multimode excitation lasers enables STORM over fields of view >100 mm, which is larger than is available from most commercial SMLM microscopes. This leads to much larger data sets and a significantly increased SMLM data processing challenge. To address this, we implemented parallelised SMLM data processing on a high-performance computing (HPC) cluster for the first time to our knowledge [6]. Using Bio-Formats, we can divide the SMLM data into subsets for parallel processing on different HPC nodes and thereby significantly reduce the data-processing time. We implemented this using ThunderSTORM but our approach should work easily with any ImageJ plug-in.
The ability to scale the data processing for large SMLM data sets is a critical step towards developing high-throughput SMLM. We are working towards this goal and have implemented easySTORM on motorised microscopes to enable imaging of arrays of samples. To date we have demonstrated automated easySTORM implemented with arrays of cells in 8-well chambers in a motorised microscope incorporating a home-built optical autofocus system [7].
Super-resolved microscopy references
- Stimulated emission depletion microscopy with a supercontinuum source and fluorescence lifetime imaging,
E. Auksorius, B. R. Boruah, C. Dunsby, P. M. P. Lanigan, G. Kennedy, M. A. A. Neil and P. M. W. French
Opt Lett 33 (2008) 113-115 - 3-D stimulated emission depletion microscopy with programmable aberration correction
M. O. Lenz, H. G. Sinclair, A. Savell, J. H. Clegg, A. C. N. Brown, D. M. Davis, C. Dunsby, M. A. A. Neil and P. M. W. French
J. Biophotonics 7 (2013) 29-36 - easySLM-STED: implementing stimulated emission depletion microscopy with straightforward alignment, aberration correction, extended field of view, and multiple beam scanning using a spatial light modulator.
F. Görlitz, S. Guldbrand, T. H. Runcorn, R. T. Murray, A. L. Jaso-Tamame, H. G. Sinclair, E. Martinez-Perez, J. R. Taylor, M. A. A. Neil*, C. Dunsby* and P. M. W. French*, Journal of Biophotonics 11 (2018) e201800087 - Mapping Molecular Function to Biological Nanostructure: Combining Structured Illumination Microscopy with Fluorescence Lifetime Imaging (SIM + FLIM)
Frederik Görlitz, David S. Corcoran, Edwin A. Garcia Castano, Birgit Leitinger, Mark A. A. Neil, Christopher Dunsby and Paul M. W. French, Photonics 4 (2017) 40 - easySTORM: a robust, lower-cost approach to localisation and TIRF microscopy
K. Kwakwa, A. Savell, T. Davies, I. Munro1 S. Parrinello, M.A. Purbhoo, C. Dunsby, M.A.A. Neil and P.M.W. French
J. Biophotonics 9 (2016) 948–957 - Accelerating single molecule localisation microscopy through parallel processing on a high-performance computing cluster HPC STORM, I. Munro*, E. García1*, M. Yan*, S. Guldbrand, S. Kumar, K. Kwakwa, C. Dunsby, M.A.A. Neil# and P.M.W. French#, J Micros. 273 (2019) 148-160
- Automated multiwell plate STORM: towards open source super-resolved high content analysis,
F. Görlitz, J. Lightley, S. Kumar, E. Garcia, M. Yan, R. Wysoczanski, Y. Alexandrov, J. R. Baker, P. J. Barnes, I. Munro, L.E. Donnelly, C. Dunsby, M.A.A. Neil and P. M. W. French
Paper 11076-4 at European Conference on Biomedical Optics, Munich 2019