A MICA award with Magnus Life Science funded by the MRC (MR/K011561/1)
Investigators
Professor Paul French (PI), Dr James McGinty - Photonics Group, Physics Department, Imperial College London
Professor Margaret Dallman, Dr Laurence Bugeon - Department of Life Sciences, Imperial College London
Dr Paul Frankel - Division of Medicine, UCL
Professor Matilda Katan - Structural Molecular Biology, UCL
Professor Simon Arridge - Computer Science, UCL
Summary
Our overarching aim is to develop a new optical technology platform to directly image mechanisms of disease throughout whole live organisms as they progress over time with a view to improving our understanding of" global" responses to disease progression, and to specific treatments, in order to develop new therapies. To visualise internal processes requires transparent organisms. Zebrafish embryos are particularly interesting because they are inherently transparent and are vertebrates - and so represent better human biology than other transparent organisms. They are increasingly used for biomedical research, with cellular processes being studied using microscopes, and many zebrafish disease models have been produced, including for cancer and inflammation. Unfortunately, the use of embryos precludes full studies of disease progression such as tumour development or chronic inflammation and it would be useful to be able to image such processes in more mature fish. Imaging larger fish, however, is challenged by the onset of absorption and scattering of light and the lack of instrumentation for high resolution 3-D imaging through cm scale samples. We would work with non-pigmented mutations of zebrafish and develop new techniques to counter light scattering that we would implement in a novel tomographic imaging system to enable detailed 3-D imaging of changes in tissue structures, cell migration and signalling processes throughout whole juvenile and adult live zebrafish.
Such "global" 3-D imaging is important for cancer because tumour cells typically spread from their original site to other organs. This is called metastasis, which is very difficult to treat and often leads to death. Observing the growth and spread of tumour cells throughout an organism and how they grow at new sites is not possible using the mammalian disease models such as mice that are often used to study cancer biology because they are not optically accessible. Global 3-D imaging is also important for inflammation to study how the response to pollutants spreads throughout an organism, leading to chronic inflammation, and how the body's defences recruit immune cells. However, even with non-pigmented zebrafish, the established optical imaging techniques are restricted to image near the surface and are strongly compromised by optical scattering. Our approach built on a method called "optical projection tomography" that is similar to x-ray tomography but works with visible light. We combined it with novel image reconstruction software and applied this novel imaging platform to the study of cancer and inflammation using genetically modified adult zebrafish. To study cancer, we bred zebrafish in which we induced tumours with a chemical reagent. These cancer models resemble the disease state in humans. To study inflammation, we used zebrafish models to study the progression of inflammation resulting from a range of treatments. We also used fluorescently labelled "biosensors" that change their fluorescence properties when specific molecular interactions take place as part of the signalling processes that determine normal cellular function and which become dysregulated in cancer and inflammation. By observing when and where signalling processes are activated or otherwise, we aim to study the effects of potential therapies such as anti-cancer or anti-inflammatory drugs.
Outcomes
During this research project we developed a new platform for global 3-D optical molecular imaging of biological processes and disease mechanisms in live embryo, juvenile and adult zebrafish, based on optical projection tomography (OPT) and fluorescence lifetime imaging (FLIM). OPT is the optical analogue of x-ray computed tomography and entails acquiring a series of 2-D images as the sample is rotated and then reconstructing a 3-D image from this data set of “projection images”. We developed a new OPT instrument to enable us to acquire 3-D images of live zebrafish, in particular applying OPT to adult zebrafish for the first time1. The capability to image zebrafish after 15 days post fertilization (dpf) is important because by this stage fish are developing their adaptive immune system and so provide a more complete model in which to study disease mechanisms and the response to therapies. This new capability should be important for drug discovery.
1 Quantitative in vivo optical tomography of cancer progression & vasculature development in adult zebrafish, Sunil Kumar, Nicola Lockwood, Marie-Christine Ramel, Teresa Correia, Matthew Ellis, Yuriy Alexandrov, Natalie Andrews, Rachel Patel, Laurence Bugeon, Margaret J. Dallman, Sebastian Brandner, Simon Arridge, Matilda Katan, James McGinty,*, Paul Frankel,*, Paul M. W. French*, Advance preview in Oncotarget: http://www.impactjournals.com/oncotarget/index.php?journal=oncotarget&page=article&op=view&path[]=9756&author-preview=7j0
Outcomes
- Compressive Sensing OPT
- Multiplexed OPT for in vivo imaging of adult zebrafish
- Data work flow for OPT
- Longitudinal imaging of zebrafish using OPT
- In vivo CS-OPT of liver cancer disease model in adult zebrafish
- In vivo FLIM OPT of FRET biosensors
- Transgenic zebrafish lines
- 3-D cell tracking during in vivo OPT acquisitions
- Remote focal scanning OPT
Traditionally CT and OPT require 100’s of projection images, which would normally extend the anaesthetic time required to unfeasible periods for in vivo imaging of zebrafish and so we developed a new “compressive sensing” approach based acquiring only 10’s of projection images and using an iterative computational technique to reconstruct the 3-D images2. Compressive sensing OPT thus permits faster image acquisition, which is important for in vivo studies.
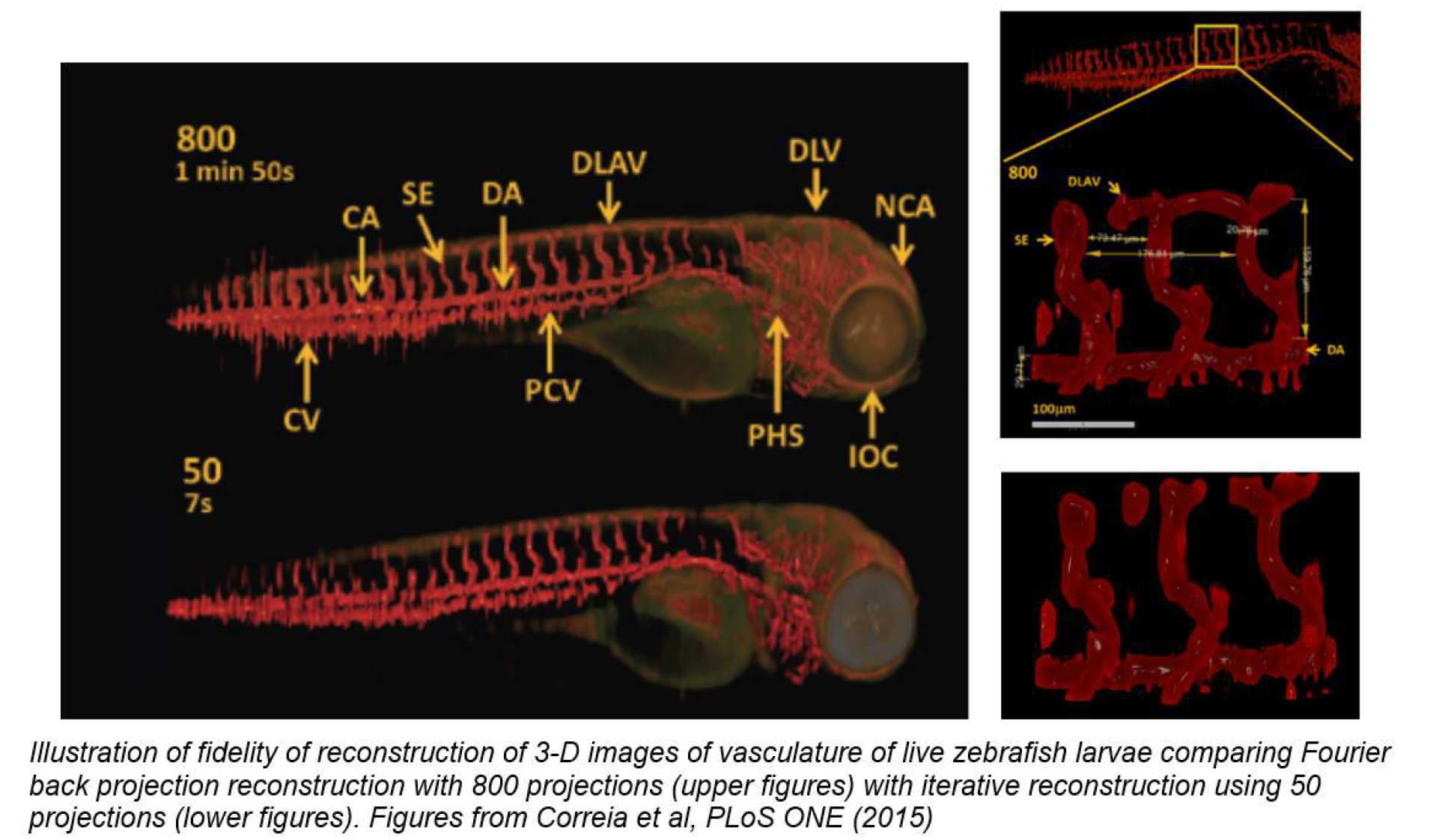
2 Accelerated Optical Projection Tomography Applied to In Vivo Imaging of Zebrafish”, T. Correia 1, N. Lockwood, S. Kumar, J. Yin, M-C Ramel, N. Andrews, M. Katan, L. Bugeon, M J Dallman, J McGinty*, P. Frankel*, P. M W French* and S. Arridge*, PLoS ONE 10(8): e0136213, (2015) doi:10.1371/journal.pone.0136213
For imaging live adult zebrafish, we implemented our novel optical configuration with two multiplexed imaging arms that act together to increase the light collection efficiency and the spatial resolution of the system3. This provides the capability to acquire full 3-D images of adult zebrafish labelled with eGFP and mCherry with a field of view of 3 cm, requiring acquisition times on the order of a minute per spectral channel. The instrument incudes a custom designed 8 window chamber with temperature control to allow further multiplexed image acquisition of transmitted and scattered light as well as fluorescence. It can also be configured for 3-D FLIM.
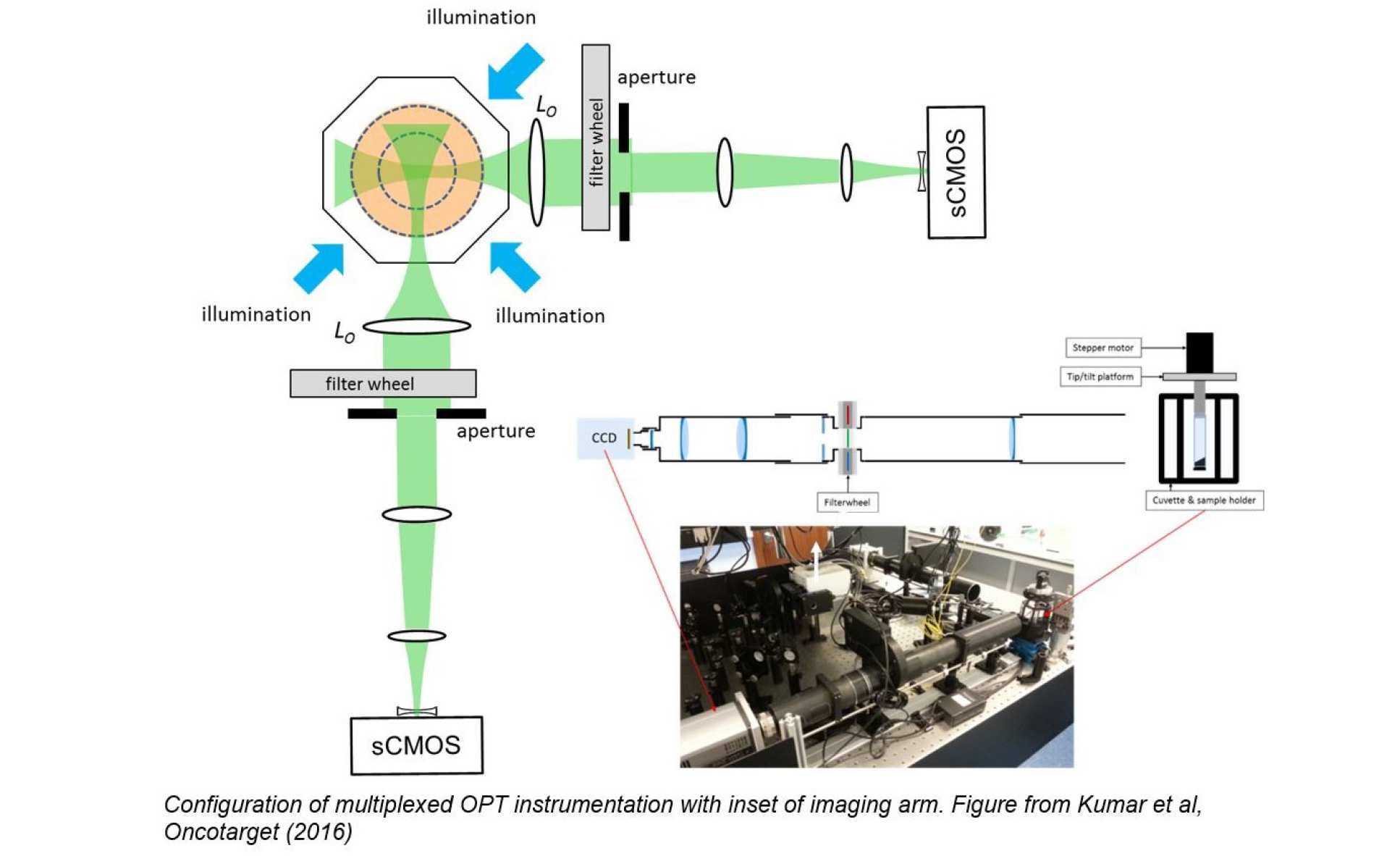
3 Simultaneous angular multiplexing optical projection tomography at shifted focal planes, L. Chen. J. McGinty and P. M. W. French, Opt Lett 38, (2013) 851; doi: 10.1364/OL.38.000851
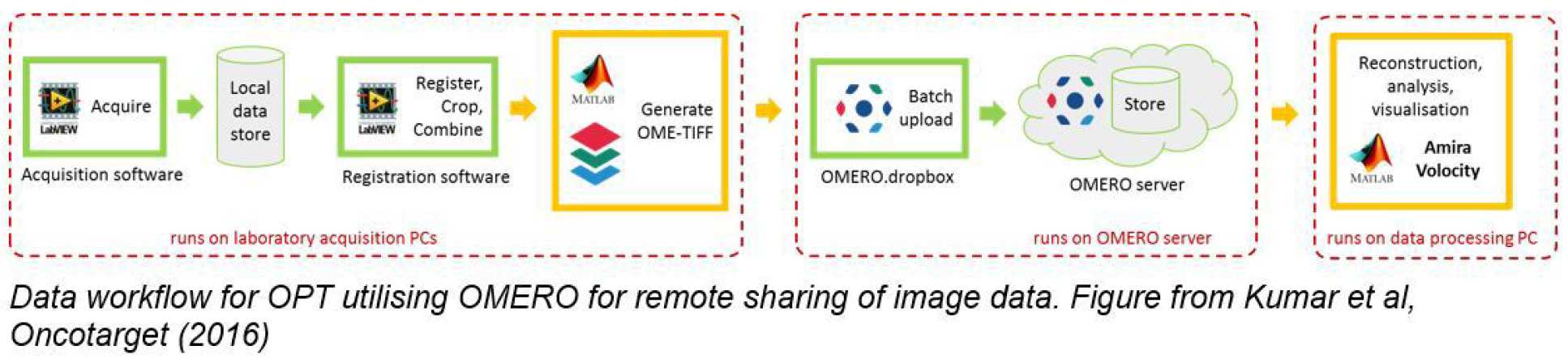
Besides the challenges associated with generating, maintaining and imaging the zebrafish lines we have also had to address the challenges associated with the data management of this OPT data. To this end we have implemented an OMERO-based solution that stores “corrected raw image data” rather than rendered 3-D stacks since the former are typically ~1 Gb/channel while the latter are >240 Gb/channel. Accordingly, we have developed a semi-automatic registration programme that co-registers the two image stacks from the multiplexed cameras and writes OME-TIFF files that can be automatically uploaded to an OMERO server. We then download OPT image data sets on demand and can reconstruct the 3-D volumetric images “on-the-fly” using fast reconstruction programmes implemented in a GPU incorporated into a desktop work station. Alternatively, we use the iterative TwIST algorithm to reconstruct the undersampled CS-OPT data sets. This OMERO-based approach lets us share data between colleagues in different buildings/institutions and is probably the most practical means to make data publically accessible.
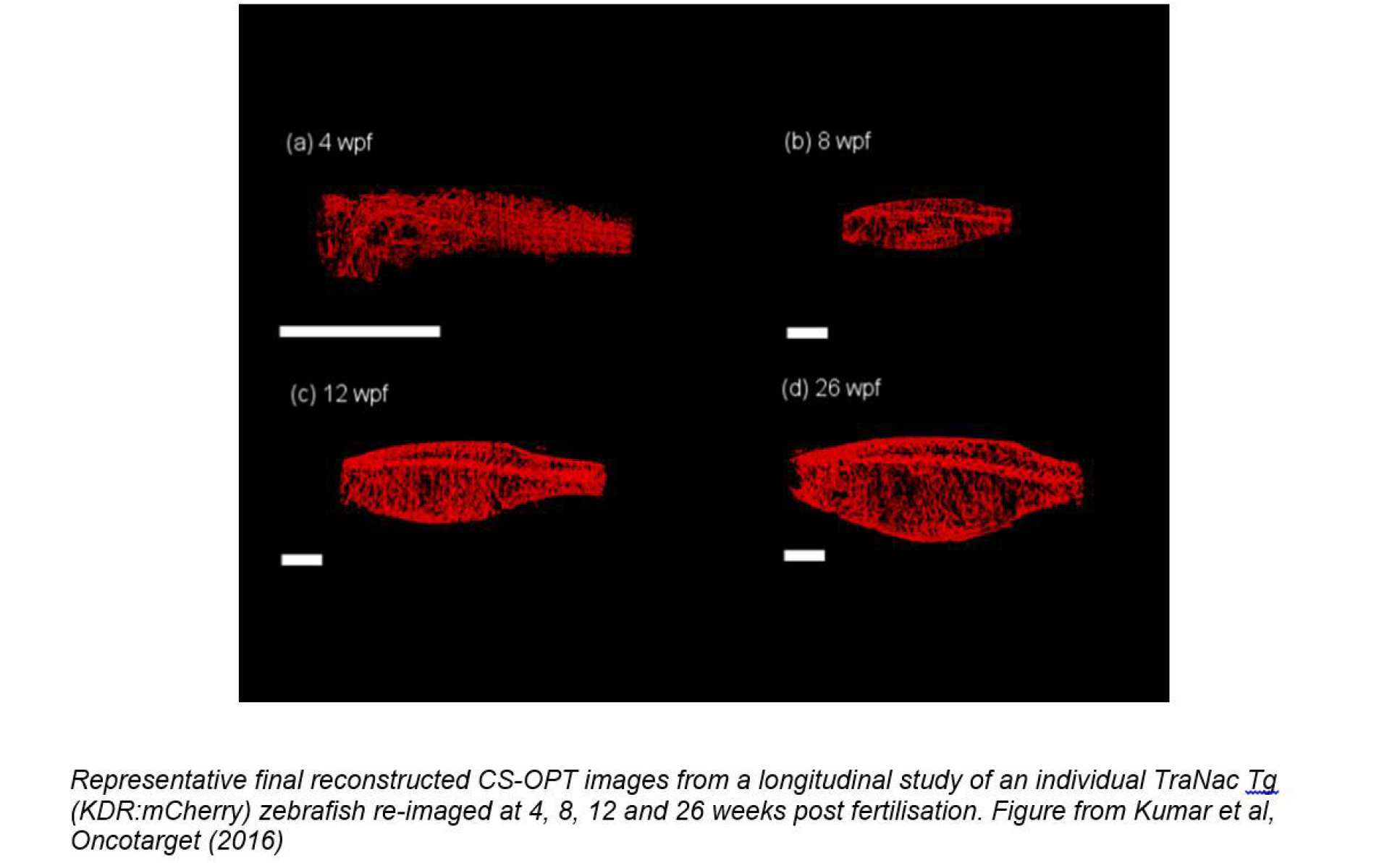
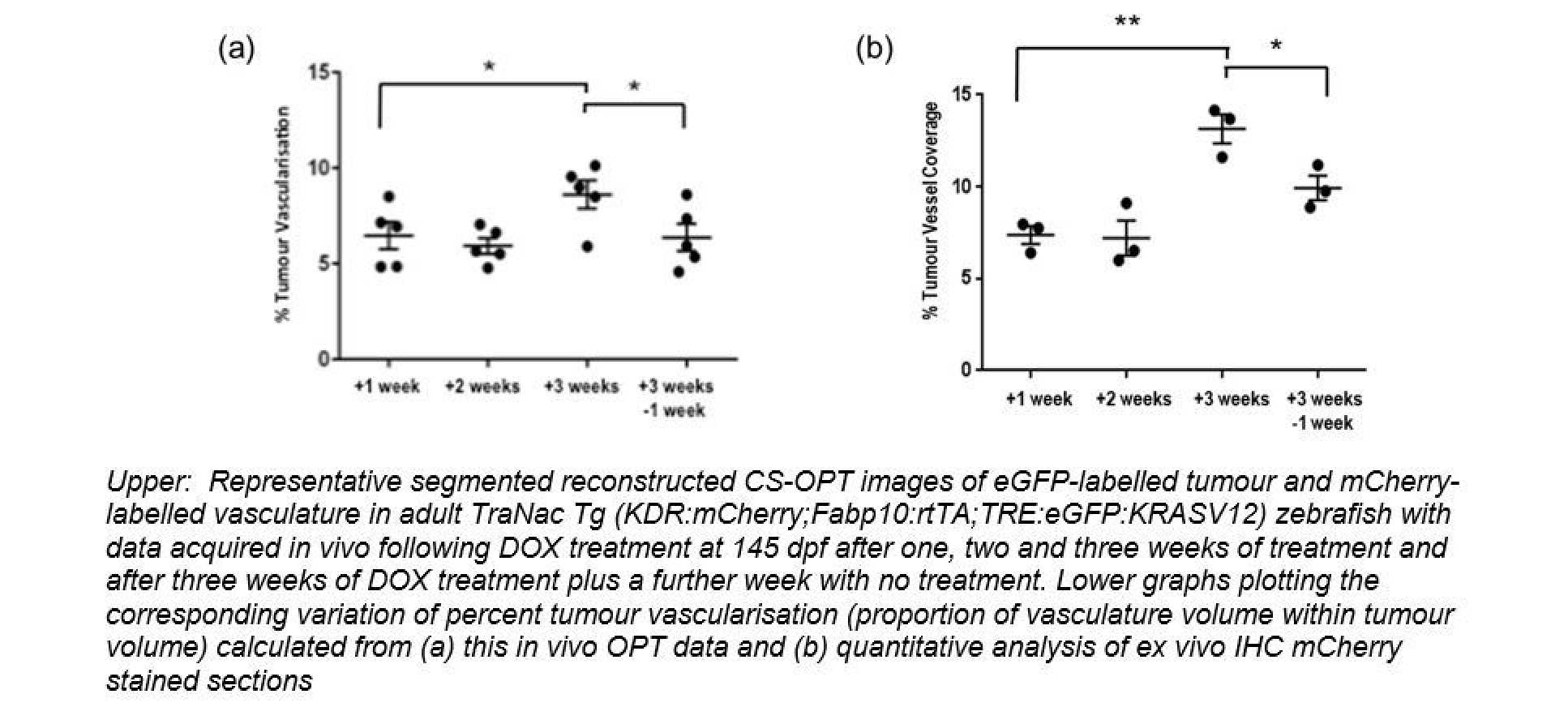
OPT can be used with transmitted light or with fluorescence and the low illumination intensities associated with the wide-field illumination of OPT present minimal phototoxicity. This enables the same fish us to be repeatedly imaged over time, thereby enabling longitudinal studies. This is important because it can provide more useful data than studies that require fish to be sacrificed at different endpoints and it can reduce the number of fish required for drug testing or other studies. In this project we imaged transgenic zebrafish lines that have been genetically engineered to suppress pigmentation (making them more transparent) and express fluorescent proteins to provide specific molecular contrast. The figure below shows images acquired over 26 weeks post fertilisation (wpf) from a longitudinal study of the same zebrafish expressing mCherry to label the vasculature.
In this project our key aim was to explore the use of OPT of zebrafish as a new preclinical modality to study disease mechanisms with a view to applications in drug discovery. To establish and disease model for cancer we generated a zebrafish line expressing mCherryFP labelling vasculature (blood vessels) and eGFP labelling a liver tumour than can be chemically induced. We have shown that we can acquire in vivo 3-D images of the vasculature and tumour in zebrafish aged more than 100 dpf and have undertaken a cross-sectional study where we demonstrated that the 3-D image data we obtain from in vivo OPT correlates with data obtained using standard histopathology techniques.
We have also implemented FLIM combined with OPT in set-ups for imaging larvae and adult zebrafish and. FLIM entails exciting a fluorescent sample with a short (picosecond) optical pulse and using ultrafast time-resovled detectors to measure how fast the resulting fluorescence emission signal decays following excitation. This “fluorescence lifetime” can tell us about the local molecular state of the fluorescent molecules. In particular, if two proteins of interest are labelled with appropriate fluorescent molecules (fluorophores), then their close proximity can lead to a transfer of energy between the fluorophores that is called “Förster resonant energy transfer” (FRET). The fluorescence lifetime of the molecule from which the energy is transferred (the “donor”) decreases when FRET occurs and this can be exploited as a robust readout of protein interactions. This phenomenon can be utilised in FRET biosensors, which are molecular constructs labelled with both “donor and “acceptor” fluorophores that change their configuration when they bind their target analyte, which can be a signalling molecule. If the change in configuration leads to a change in the distance between the donor and acceptor fluorophores, then mapping the donor fluorescence lifetime can effectively provide a map of the signalling molecules. In this project we have developed FLIM OPT to map the 3-D distribution of signalling molecules in live zebrafish. We have therefore developed a range of transgenic zebrafish lines that genetically express FRET biosensors to read out signalling molecules associated with apoptosis, (cell death), inflammation and cancer.
To date we have made the most progress mapping apoptosis in live zebrafish embryos using a FRET sensor for Caspase 3, which is a signalling molecule associated with apoptosis4. We have shown that apoptosis induced by gamma irradiation, can be mapped in zebrafish larvae expressing a Caspase 3 FRET biosensor using in vivo 3-D FLIM.
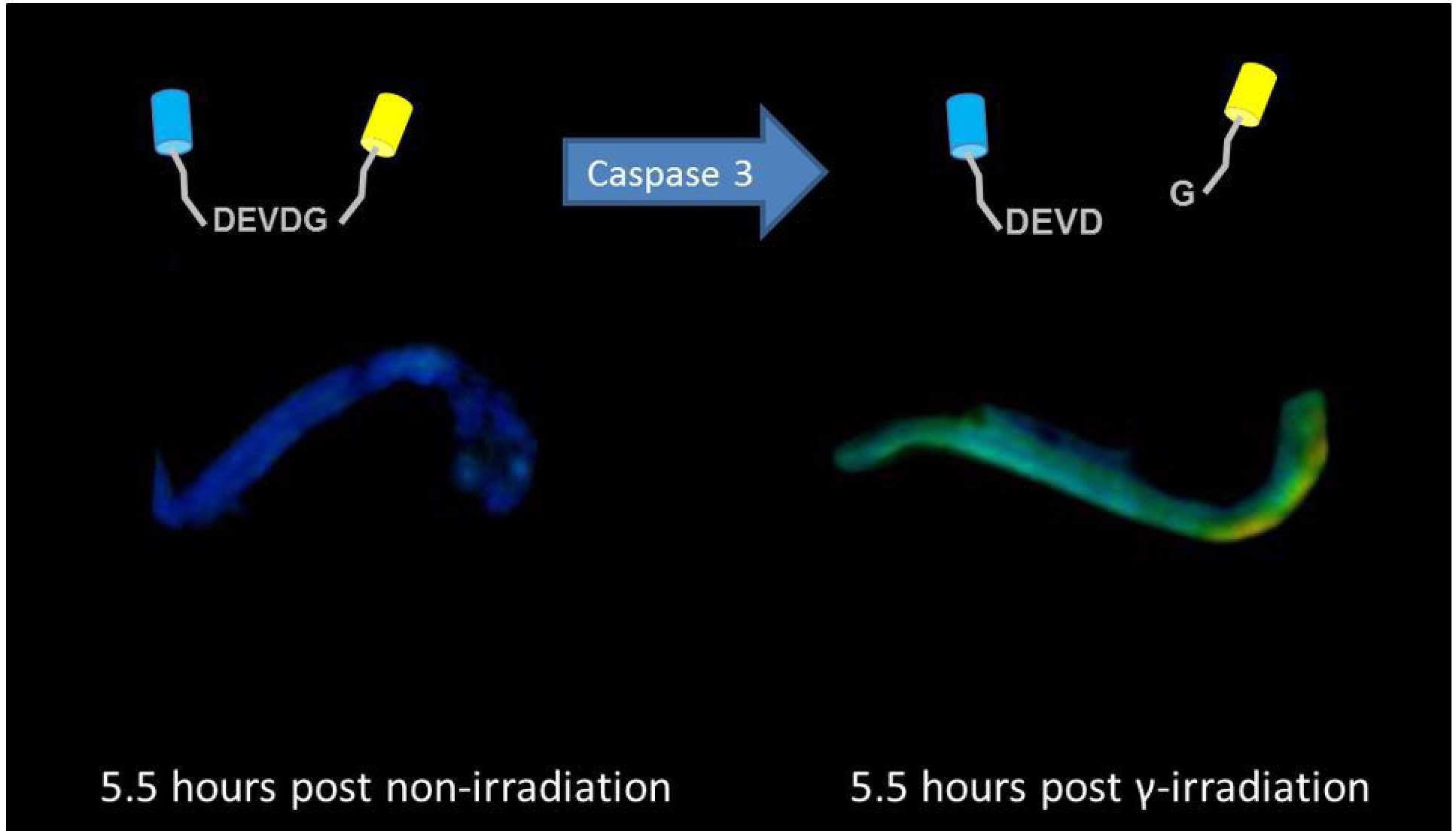
4 Visualising apoptosis in live zebrafish using fluorescence lifetime imaging with optical projection tomography to map FRET biosensor activity in space and time, N. Andrews, M-C Ramel, S. Kumar, Y. Alexandrov, D. J. Kelly, S. C. Warren, L. Kerry, N. Lockwood, A. Frolov, P. Frankel, L. Bugeon, J. McGinty, M. J. Dallman* and P. M. W. French*, Early view J. Biophotonics 1–11 (2016) / DOI 10.1002/jbio.201500258
|
TraNac TG(fabp10-rtTA) TraNac TG(TRE-Src bios) TraNac TG(TRE-Rac1 bios) TraNac TG(TRE-MT1-MMP) TraNac TG(TRE-iRFP-KRasV12) TraNac Tg(Ubi:Rac1 bios) TraNac Tg(Ubi:SECFP) |
TraNac Tg(Ubi:Caspase1 YVHDA bios) TraNac Tg(Ubi:Caspase3 bios) TraNac Tg(MPEG1:Caspase1 YVHDA bios) TraNac Tg(MPEG1:Caspase3 bios) TraNac Tg(IFABP 4.5Kb:Caspase1 bios) TraNac Tg(MPEG1:Caspase1 bios; KDR:mCherry) TraNac Tg(LysC:Caspase3 bios) TraNac Tg(LysC:Caspase1 bios) |
During this project we also demonstrated a novel approach to 3-D cell tracking during OPT acquisitions with multiplexed image acquisition that essentially entails focussing two imaging channels to the same depth in the sample during an OPT acquisition and localising features in 3-D using triangulation at each time point, i.e. each image acquisition. Thus, while the full OPT acquisition may take 100’s of seconds to acquire, dynamic features such as cells can be tracked with a time resolution equal to the frame rate of the cameras – which can be 100’s frames/second – although in practice integration times have to be sufficiently long to acquire images with adequate signal to noise ratio. The proof of principle was demonstrated using GFP-labelled neutrophils in live zebrafish embryos as illustrated below where the transmission OPT image of a live 2 days post fertilisation (dpf) zebrafish embryo tail is shown overlaid with tracks indicating the trajectories of the neutrophils over ~25 minutes following a tail wound, colour-coded by position and velocity5.
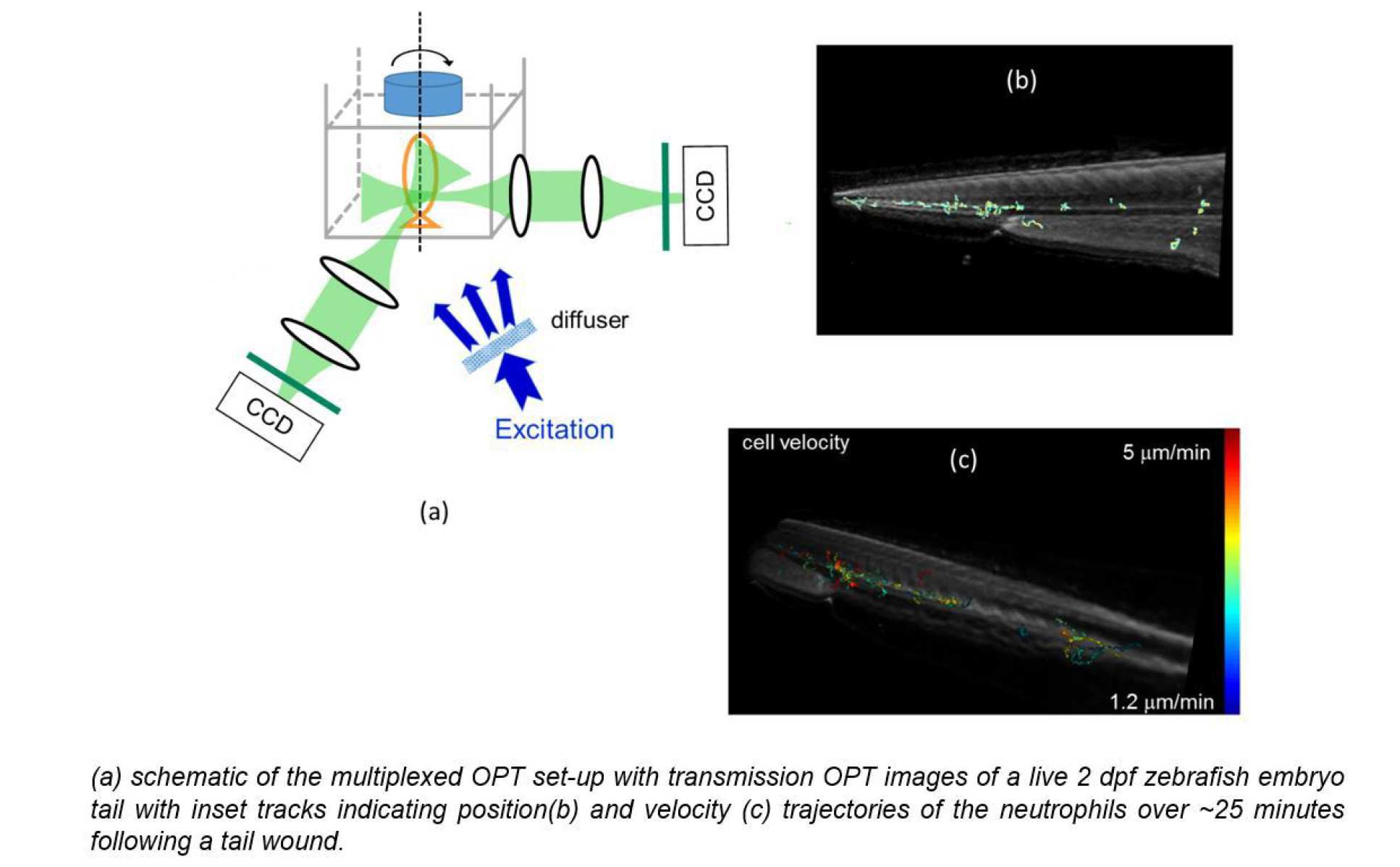
5 Mesoscopic in vivo 3-D tracking of sparse cell populations using angular multiplexed optical projection tomography, L Chen, Yuriy A, Sunil Kumar, N Andrews, M J. Dallman, P M. W. French* and J McGinty*, Biomedical Optics Express 6 (2015) 1253- 1261, DOI:10.1364/BOE.6.001253
To extending the sample size while preserving spatial resolution of OPT, we have explored a focal scanning approach implemented with an electrically tunable lens6 as an alternative to multiplexed imaging arms. By rapidly scanning the imaging lens during the acquisition of each projection image, an “in focus” image is recorded throughout the sample while the out of focus background is removed through the usual Fourier back projection reconstruction process.
6 Remote focal scanning optical projection tomography with an electrically tunable lens, L Chen, S. Kumar, D. Kelly, N. Andrews, M.J. Dallman, P. M W French and J. McGinty, Biomed Optical Express, 5 (2014) 3367-3375 DOI:10.1364/BOE.5.003367