Contact
Dr Tamas Korcsmaros
10th Floor,Commonwealth Building,Hammersmith Campus.
Group website: http://KorcsmarosLab.org
X (Twitter): @Korcsmaroslab
LinkedIn: Tamas Korcsmaros
What we do
The Korcsmaros Group combines computational approaches (network biology and AI) and experimental technologies (organoids, organoids-on-chip and multi-omics) to study complex systems in the gut. Our goal is to understand biological systems related to gut homeostasis and to facilitate precision medicine and personalised microbial therapies in the field of inflammatory bowel disease (IBD). Our vision is that with the combination of systems biology and precision medicine resources, we are going to understand better the patient-specific nature of host-microbe interactions in the gut, and identify effective therapeutical options preventing the onset of IBD or relapse. To achieve this vision, our aim is to work on the multi-omics analysis of gut organoids to create computational network models of key intestinal cells interacting with other host cells or with microbial products, and then validate the predictions in a patient-specific organoid testing system.
Why it is important
The intestinal epithelium in the gut often does not work properly. The example we focus on is Inflammatory Bowel Disease (IBD), a chronic pathology of the gut that affects nearly 10 million patients worldwide (from which 500,000 patients are in the UK) and costs the NHS >£1.8 billion annually. Therefore, there is a strong need to further understand the pathological mechanisms in IBD and to personalise treatments for individual patients. The group is interested in finding ways to improve how the epithelial layer works and how we can best match the efficient treatments with those patients who respond to them. By building computational network models we can predict the metabolites and microbial connections to epithelial cells in healthy versus diseased states which can be tested experimentally using gut organoids as a model system. Organoids are a powerful and promising tool to investigate the physiological and cell-specific effects of diseases such as IBD.
How it can benefit patients
Inflammatory bowel disease (IBD) is a ‘patient-specific’ disease with different treatment plans required for different patients. Remission rates for IBD remain low demonstrating a limit on how effective treatments are for patients. We use computational network modelling and experimental organoid-based systems to better understand patient-specific pathomechanisms and IBD or remission associated host-microbe interactions. This can help to identify effective therapeutic options and microbial supplementation for a specific IBD patient.
Summary of current research
- Network Medicine Approaches in IBD
- Multi-omics Analysis of Intestinal Organoids
- Analysing Host-Microbiome Interactions
- Mapping Autophagy regulation in the Gut
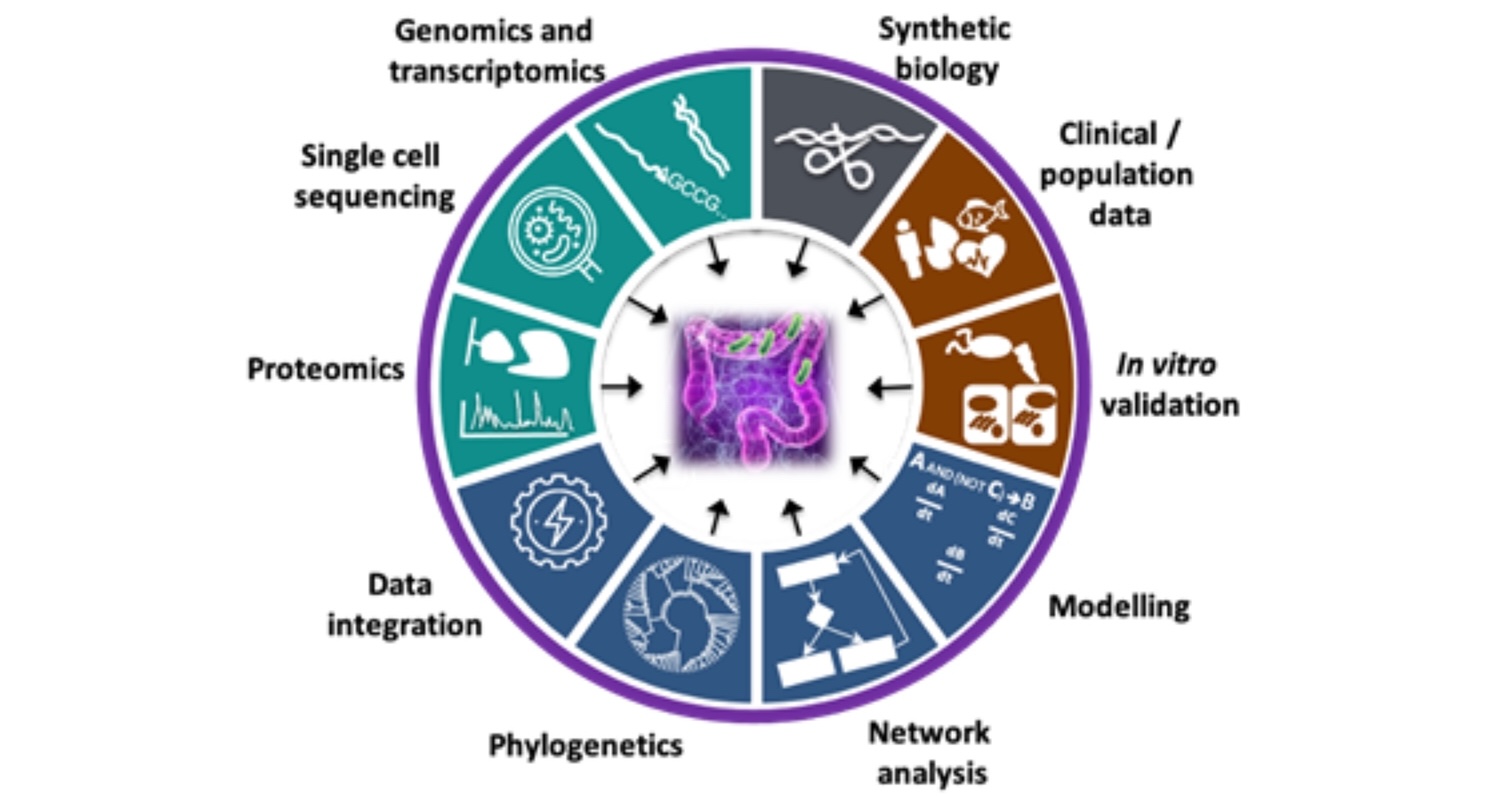
Inflammatory Bowel Disease (IBD) is a disorder of the human gastrointestinal tract characterised by inflammation of the gut mucosal layer and dysbiosis of the microbiome. IBD can broadly be classified into two diseases - Ulcerative Colitis (UC) and Crohn’s Disease (CD) - which are increasingly prevalent in the global population. However, IBD is a complex disease influenced by a variety of extrinsic and intrinsic factors such as genetics, microbiome, lifestyle (diet, smoking etc), xenobiotic exposures etc. In the Korcsmaros group, we aim to understand IBD using multiple different angles and various omics techniques as input data sources. To investigate patient-specific pathogenesis in IBD, we collaborate with Séverine Vermeire’s IBD team from Leuven and with the UK IBD Genetic Consortia. We use patient-specific genomics and transcriptomics information from these collaborators to decipher how patient-specific genomic background contributes to IBD.
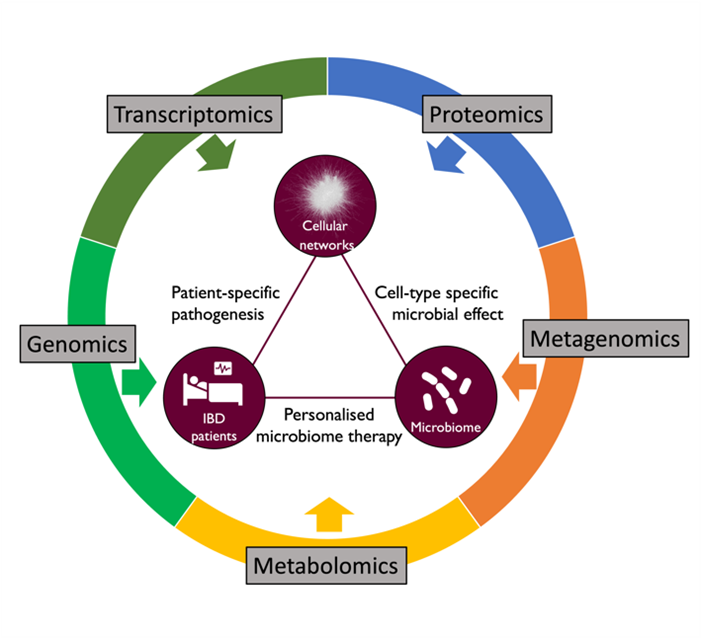
Figure 1: Our integrative research approaches to study IBD
In healthy individuals, the gastrointestinal tract relies on highly sophisticated intercellular interactions to sense, adapt and respond appropriately to its cells’ immediate and distant environments. These include not only external factors (e.g. diet, resident, transitory probiotic or infectious microbes) but also other host cells from near or far distances (e.g. the underlying mesenchyme, the gut-associated innate and adaptive immune cells, the enteric nervous system, or even distant organs like the liver, the lung or the brain). Dysbiosis of the microbiota or any alteration of host cell-to-cell crosstalk can result in complex multifactorial pathologies, including Inflammatory Bowel Disease (IBD). We are combining in silico systems biology approaches with experimental data generation and validation using the human colon organoid model (colonoids). Organoids are 3D structures, grown from stem cells that recapitulate the normal physiology and the different cell types of the original organ. Intestinal organoids enable the in vitro study of the regulation/dysregulation of epithelial homeostatic functions, cell-to-cell interactions and host-microbiome interactions happening in the gut during health and disease. Furthermore, the generation of multi-omics data from organoids, in combination with the application of in silico network tools, such as our OmniPath database, enables the generation of molecular interaction networks allowing contextual and multiscale analysis of the ‘omics data. These approaches facilitate deciphering molecular mechanisms involved in gut homeostasis. By increasing the understanding of the underlying factors altered in intestinal dysbiosis-associated pathologies and the beneficial effects commensal bacteria may have on intestinal cell function, we hope to pave the way for translational developments in the prevention and treatment of gastrointestinal disorders.
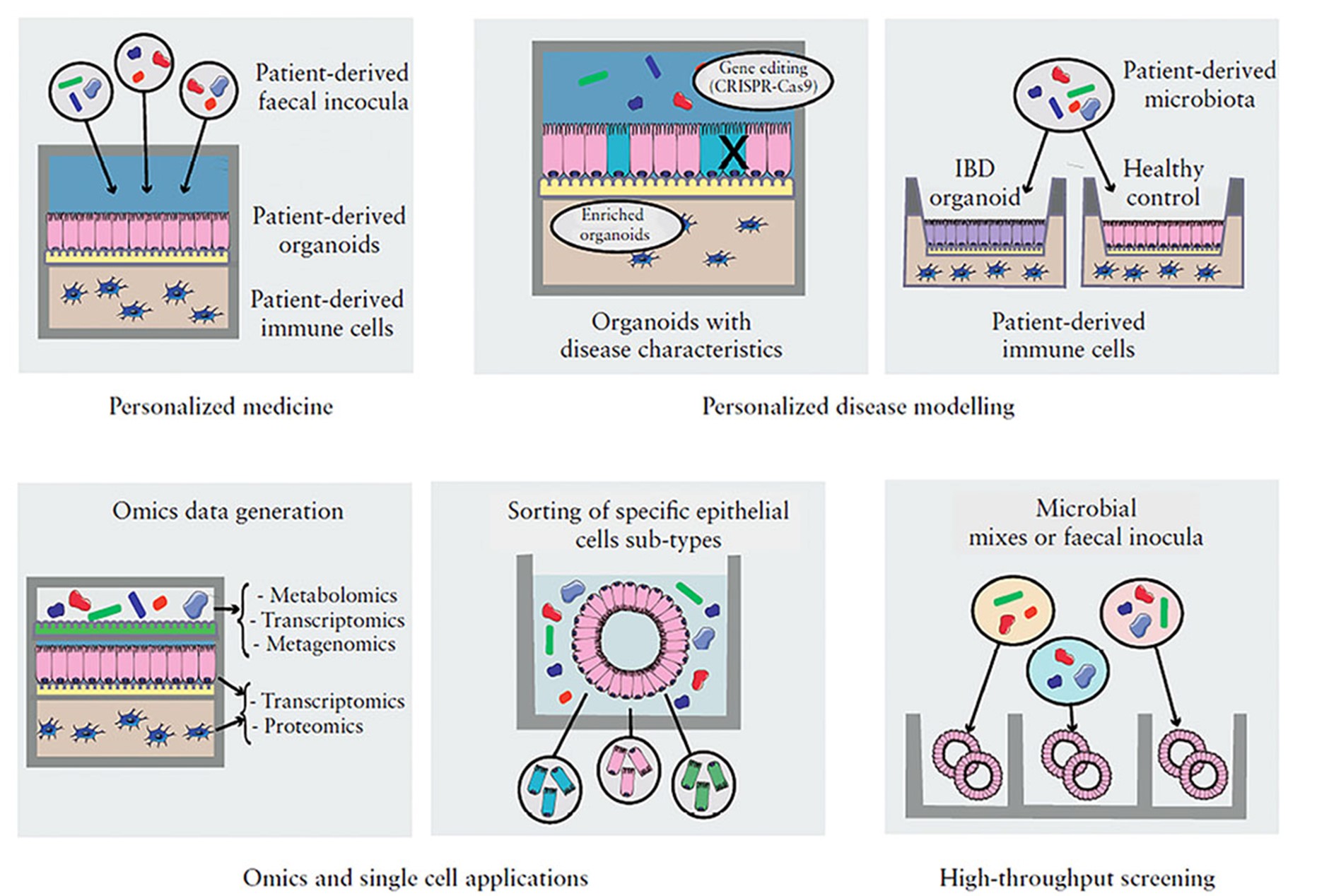
Figure 2: Organoid-based model applications
The microbiome plays a vital role in maintaining host systems' homeostatic (healthy) functions. Disrupting the healthy microbiota - by external (temperature, hygiene, etc.) or internal factors (inflammation, ageing, etc.) - causes a dysbiotic (unhealthy) state. Disruption of the microbial equilibrium between commensal and pathogen species could lead to the development of various disorders. Though differences in the microbiota between healthy and unhealthy phenotypes have been reported, the mechanistic role of the microbiota in modulating homeostatic processes remains largely unexplored.
As a focused, gap-filling attempt to address this problem, we have developed in silico pipelines to predict host-microbe interactions and analyse the effect of microbes on human signalling pathways based on omics data using network biology approaches. In industrial collaboration with Unilever, we have been testing and fine-tuning our workflows on data from the gastrointestinal tract.
Autophagy is a common recycling process in which cells degrade their unnecessary or damaged parts. It plays an important role in intestinal homeostasis, and when it is not properly functioning, it can lead to gastrointestinal diseases such as Inflammatory Bowel Disease (IBD). In our lab, we are interested in studying key regulators of autophagy in different epithelial cell types in the gut in both health and disease states. This understanding may shed light on the role of autophagy in specific cell types during the development of gut diseases, and potentially lead to new targets for drug purposes. Additionally, we are interested in how autophagy in the gut is modulated by neighbouring cells or commensal bacteria, and how these cell-cell interactions are disrupted during disease. To achieve our goals, we use a combination of experimental and computational approaches. In particular, we use “gut organoids” (a 3D miniature version of the human gut) from healthy or IBD patients to study autophagy regulation in different cell types and assess autophagy modulation by external factors such as microbial compounds. Additionally, we combine single-cell RNA sequencing (scRNA-seq) technologies with previously developed bioinformatics resources such as the AutophagyNet to reconstruct and analyse the intracellular and intercellular signalling related to autophagy in the gut.
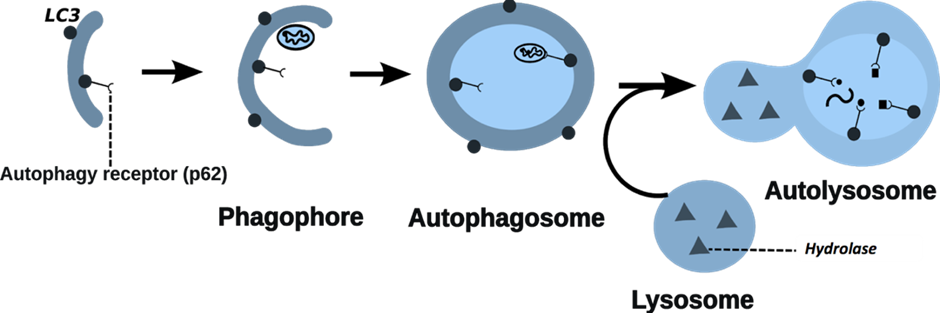
Figure 3: Process of autophagy
Information
Funders
Related centres
Internal
- Dr Dezso Modos (Department of Metabolism, Digestion and Reproduction, Imperial)
- Dr Rachael Barry (Department of Metabolism, Digestion and Reproduction, Imperial)
- Prof Nick Powell (Department of Metabolism, Digestion and Reproduction, Imperial)
- Prof Horace Williams (Department of Metabolism, Digestion and Reproduction, Imperial)
- Prof Gary Frost (Department of Metabolism, Digestion and Reproduction, Imperial)
- Prof Marc-Emmanuel Dumas (Department of Metabolism, Digestion and Reproduction, Imperial)
- Prof Clare Lloyd (National Heart and Lung Institute, Imperial)
- Prof Petter Brodin (Department of Immunology and Inflammation, Imperial)
External
- Dr Julio Saez-Rodriguez (Head of Research, European Bioinformatics Institute)
- Dr Séverine Vermeire (IBD group, KU Leuven, Belgium)
- Dr Bram Verstockt (IBD group, KU Leuven, Belgium)
- Paul Wilmes group (University of Luxembourg)
- Lindsay Hall Lab (University of Birmingham, UK)
- Orsolya Kapuy (Semmelweis University, Hungary)
- Ioannis Nezis (University of Warwick, UK)
- Tom Wileman (Quadram Institute, Norwich, UK)
- Simon Carding (Quadram Institute, Norwich, UK)
- Falk Hildebrand (Quadram Institute, Norwich, UK)
- Gul L, Elias AJ, Tambaku T, Olbei M, Watters E, Bohar B, et al. Protocol for predicting host-microbe interactions and their downstream effect on host cells using MicrobioLink. Star Protoc. 2025;6(1).
- Zhang YR, Thomas JP, Korcsmaros T, Gul L. Integrating multi-omics to unravel host-microbiome interactions in inflammatory bowel disease. Cell Rep Med. 2024;5(9).
- Papp D, Korcsmaros T, Hautefort I. Revolutionizing immune research with organoid-based co-culture and chip systems. Clin Exp Immunol. 2024;218(1):40-54.
- Lo JW, Schroeder JH, Roberts LB, Mohamed R, Cozzetto D, Beattie G, et al. CTLA-4 expressing innate lymphoid cells modulate mucosal homeostasis in a microbiota dependent manner. Nat Commun. 2024;15(1).
- Kapuy O, Holczer M, Csabai L, Korcsmáros T. Oscillatory autophagy induction is enabled by an updated AMPK-ULK1 regulatory wiring. PLoS One. 2024;19(12). doi: 10.1371/journal.pone.0313302.
- Csabai L, Bohár B, Türei D, Prabhu S, Földvári-Nagy L, Madgwick M, et al. AutophagyNet: high-resolution data source for the analysis of autophagy and its regulation. Autophagy. 2024;20(1):188-201. doi: 10.1080/15548627.2023.2247737.
- Lo JW, Schroeder JH, Roberts LB, Mohamed R, Cozzetto D, Beattie G, et al. CTLA-4 expressing innate lymphoid cells modulate mucosal homeostasis in a microbiota dependent manner. Nat Commun. 2024;15(1):9520. doi: 10.1038/s41467-024-51719-6.
- Papp D, Korcsmaros T, Hautefort I. Revolutionizing immune research with organoid-based co-culture and chip systems. Clin Exp Immunol. 2024;218(1):40-54. doi: 10.1093/cei/uxae004.
- Zhang Y, Thomas JP, Korcsmaros T, Gul L. Integrating multi-omics to unravel host-microbiome interactions in inflammatory bowel disease. Cell Rep Med. 2024;5(9):101738. doi: 10.1016/j.xcrm.2024.101738.
- Alexander JL, Mullish BH, Danckert NP, Liu Z, Olbei ML, Saifuddin A, et al. The gut microbiota and metabolome are associated with diminished COVID-19 vaccine-induced antibody responses in immunosuppressed inflammatory bowel disease patients. EBioMedicine. 2023;88:104430. doi: 10.1016/j.ebiom.2022.104430.
- Bernuzzi F, Maertens A, Saha S, Troncoso-Rey P, Ludwig T, Hiller K, et al. Sulforaphane rewires central metabolism to support antioxidant response and achieve glucose homeostasis. Redox Biol. 2023;67:102878. doi: 10.1016/j.redox.2023.102878.
- Corpas M, de Mendoza C, Moreno-Torres V, Pintos I, Seoane P, Perkins JR, et al. Genetic signature detected in T cell receptors from patients with severe COVID-19. iScience. 2023;26(10):107735. doi: 10.1016/j.isci.2023.107735.
- Genna A, Alter J, Poletti M, Meirson T, Sneh T, Gendler M, et al. FAK family proteins regulate in vivo breast cancer metastasis via distinct mechanisms. bioRxiv [Preprint]. 2023 Oct 29:2023.10.27.564212. doi: 10.1101/2023.10.27.564212.
- Hajdú B, Csabai L, Márton M, Holczer M, Korcsmáros T, Kapuy O. Oscillation of autophagy induction under cellular stress and what lies behind it, a systems biology study. Int J Mol Sci. 2023;24(8):7671. doi: 10.3390/ijms24087671.
- Keller-Pintér A, Korcsmáros T, Vellai T. Managing type 2 diabetes: targeting a microbial enzyme as a novel treatment option. Signal Transduct Target Ther. 2023;8(1):444. doi: 10.1038/s41392-023-01694-z.
- Lo JW, Cozzetto D, Alexander JL, Danckert NP, Madgwick M, Knox N, et al. Immune checkpoint inhibitor-induced colitis is mediated by polyfunctional lymphocytes and is dependent on an IL23/IFNγ axis. Nat Commun. 2023;14(1):6719. doi: 10.1038/s41467-023-41798-2.
- Lobentanzer S, Aloy P, Baumbach J, Bohar B, Carey VJ, Charoentong P, et al. Democratizing knowledge representation with BioCypher. Nat Biotechnol. 2023;41(8):1056-1059. doi: 10.1038/s41587-023-01848-y.
- Ray N, Park SJ, Jung H, Kim J, Korcsmaros T, Moon Y. Stress-responsive Gdf15 counteracts renointestinal toxicity via autophagic and microbiota reprogramming. Commun Biol. 2023;6(1):602. doi: 10.1038/s42003-023-04965-1.
- Brooks-Warburton J, Modos D, Sudhakar P, Madgwick M, Thomas JP, Bohar B, et al. A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis. Nat Commun. 2022;13(1):2299. doi: 10.1038/s41467-022-29998-8.
- Csabai L, Fazekas D, Kadlecsik T, Szalay-Bekő M, Bohár B, Madgwick M, et al. SignaLink3: a multi-layered resource to uncover tissue-specific signaling networks. Nucleic Acids Res. 2022;50(D1):D701-D709. doi: 10.1093/nar/gkab909.
- Demeter A, Jacomin AC, Gul L, Lister A, Lipscombe J, Invernizzi R, et al. Computational prediction and experimental validation of Salmonella Typhimurium SopE-mediated fine-tuning of autophagy in intestinal epithelial cells. Front Cell Infect Microbiol. 2022;12:834895. doi: 10.3389/fcimb.2022.834895.
- Dr John Thomas
- Dr Rohan Sundramoorthi
- Dr Robin Brown
- Edvishka Dias
- Toby Lawrence
- Lena Weidert
- Luca Csabai
Our researchers
Dr Tamas Korcsmaros
/prod01/channel_3/media/images/people-list/t.korcsmaros.jpg)
Dr Tamas Korcsmaros
Principal Investigator
Yufan Liu
/prod01/channel_3/media/images/people-list/Yufan-Liu-photo-(002).jpg)
Yufan Liu
Research Postgraduate
Dr Marton Olbei
/prod01/channel_3/media/images/people-list/Marton-Olbei.jpg)
Dr Marton Olbei
Research Postgraduate
Inez Roegiers
/prod01/channel_3/media/images/people-list/Picture_IR.png)
Inez Roegiers
Research Postgraduate
Balazs Bohar
/prod01/channel_3/media/images/people-list/Balazs-Bohar-photo-.jpg)
Balazs Bohar
Research Postgraduate
Lejla Gul
/prod01/channel_3/media/images/people-list/Holding-PNG--tojpeg_1564655919889_x2.jpg)
Lejla Gul
Research Postgraduate