Chemical probes pave the way for a better understanding of disease development
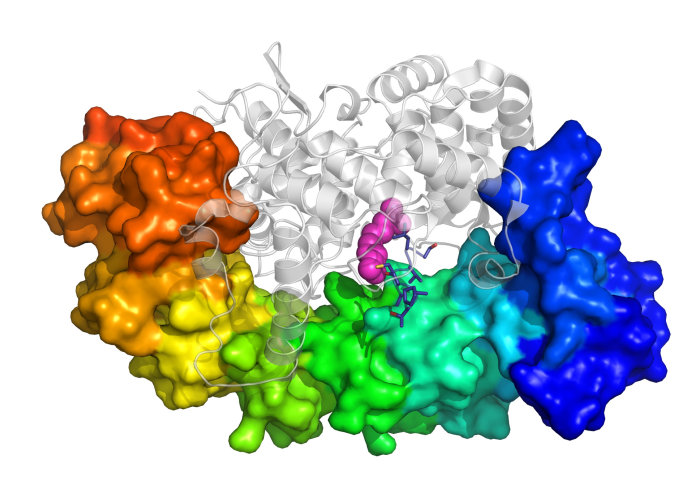
Researchers can now tag proteins in live cells that have modifications associated with disease.
Proteins produced in cells often undergo modifications by enzymes after they are formed. One type of modification, called prenylation, adds ‘tags’ to proteins that tells them where to go in the cell and how to interact with other proteins.
We developed tags that not only work without statins, but can also analyse different prenylation pathways in parallel, which has not been possible before. Professor Ed Tate
However, when prenylation goes wrong, it can lead to diseases including cancer, retinal and cardiovascular diseases, and viral infections.
Because of their role in these diseases, the enzymes that cause prenylation have been targeted by potential new drugs. For example, recent work led by Dr Beata Wojciak-Stothard from the Department of Medicine at Imperial has suggested that prenylation may be an important drug target in pulmonary arterial hypertension.
However, because the mechanisms of prenylation are difficult to study, none of the potential drugs targeting it directly have so far been approved for medical use, although indirect targeting using bisphosphonate drugs is an important treatment for osteoporosis.
Understanding the interplay
Now, researchers led by Professor Ed Tate, from the Department of Chemistry at Imperial, have developed a new way to track prenylation that does not affect the normal working of a cell, allowing them to profile the full range of prenylation inside the cell.
Professor Tate said: “All previous studies used drugs called statins to suppress prenylation and favour incorporation of chemical tags, but statins have widespread impacts on cells, making those experiments very difficult to interpret. We developed tags that not only work without statins, but can also analyse different prenylation pathways in parallel, which has not been possible before.
“This also allows us to understand the interplay between the different pathways for the first time, as well as to get a lot more information about each individually since all our analytical power can be focused on one at a time.”
The recent study, published in Nature Chemistry, describes two new chemical tags that can be used to detect protein prenylation inside living cells without any need for disruption of a normal cellular environment. The new tags match closely the natural prenylation tags found in cells and are therefore used just as readily by the prenylating enzymes.
For the first time the researchers were able to detect the full range of proteins that are prenylated in cells in a single experiment and to discover new proteins that where not previously known to be prenylated.
They were also able to observe switching between prenylation pathways in response to treatment with anticancer drugs, providing the first global insights into this process, which is important for drug resistance.
Disease insights
Prenylated proteins are known to be involved in multiple pathways that are disrupted in a disease context: one example is choroideremia.
Choroideremia is a genetic disorder that leads to potential loss of eyesight. Choroideremia patients have a mutation in a gene coding for REP-1 protein, which prevents it from functioning correctly. REP-1 is involved in a pathway that adds prenylation tags on other proteins, however, without a functioning REP-1 those other proteins do not get prenylated.
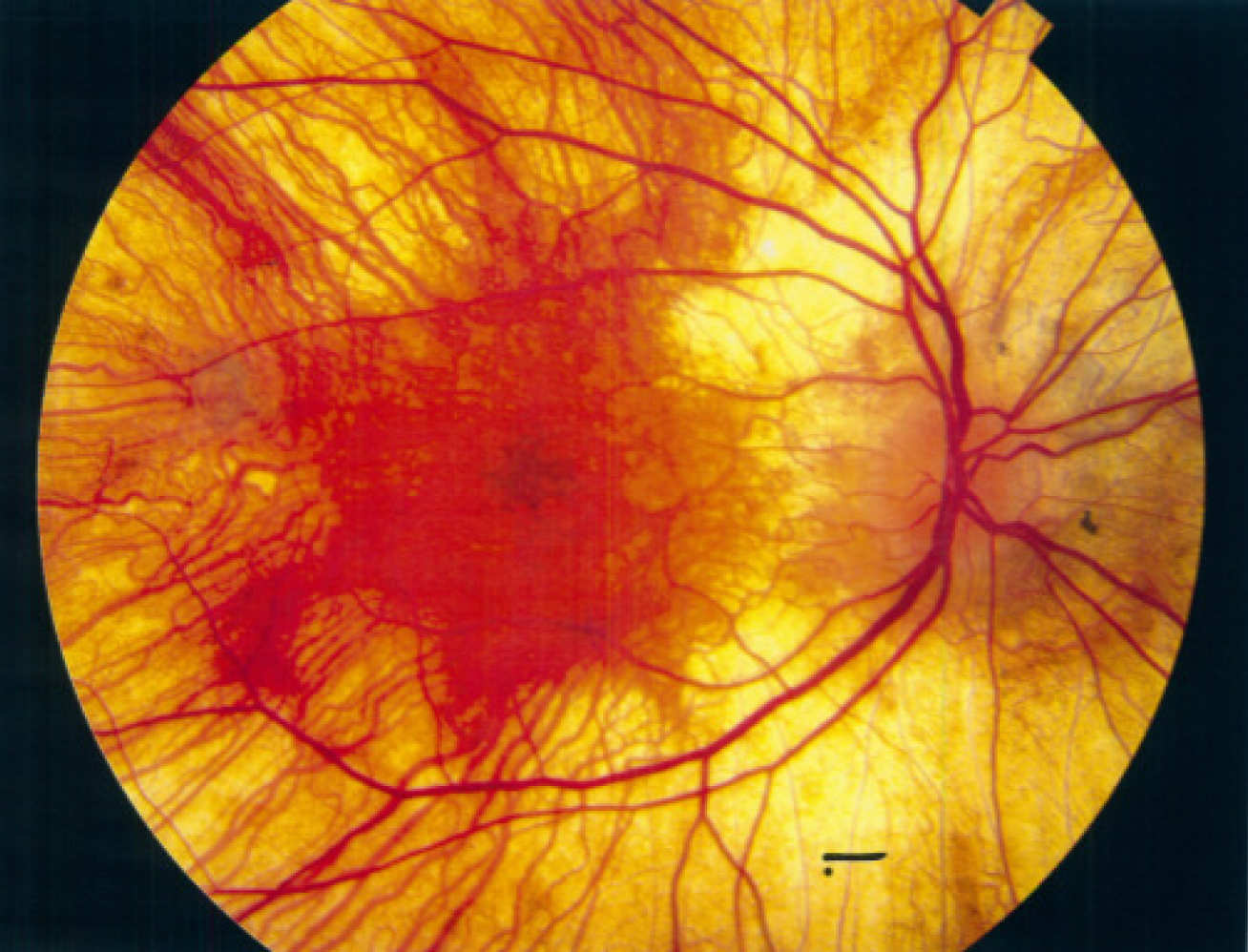
Using the newly developed chemical tags, Professor Tate’s research was able to see the global changes in prenylation patterns in a mouse model of choroideremia for the first time, and find the specific proteins that do not get prenylated due to the defective RAB-1. This insight could lead to new ways to treat choroideremia patients.
When asked what the next steps in the research using the newly developed probes will be, professor Tate said: “We would like to use them to explore in more detail how cancer cells evade prenylation inhibitors and how we might overcome this resistance.”
In addition, the research group is planning to further explore the biological functions of the newly discovered prenylated proteins.
The research was supported by Cancer Research UK, the British Heart Foundation and the European Union, and in collaboration with the groups of Beata Wojciak-Stothard (Department of Medicine at Imperial), Miguel Seabra (Universidade Nova de Lisboa) and Juan Martin-Serrano (King’s College London).
-
‘Dual chemical probes enable quantitative system-wide analysis of protein prenylation and prenylation dynamics’ by Elisabeth M. Storck, Julia Morales-Sanfrutos, Remigiusz A. Serwa, Nattawadee Panyain,Thomas Lanyon-Hogg, Tanya Tolmachova, Leandro N. Ventimiglia, Juan Martin-Serrano, Miguel C. Seabra, Beata Wojciak-Stothard and Edward W. Tate is published in Nature Chemistry.
Top image credit: Ed Tate/Imperial College London
Eye image: Michael R. Christensen, MS-IV, Virginia Commonwealth University School of Medicine; Charles Calvo, MD
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Bernadeta Dadonaite
Centre for Languages, Culture and Communication