Antiviral metabolite identified as potential serum biomarker for viral infection

A new paper from Imperial scientists has identified an antiviral metabolite as a potential serum biomarker for viral infections, including SARS-CoV-2.
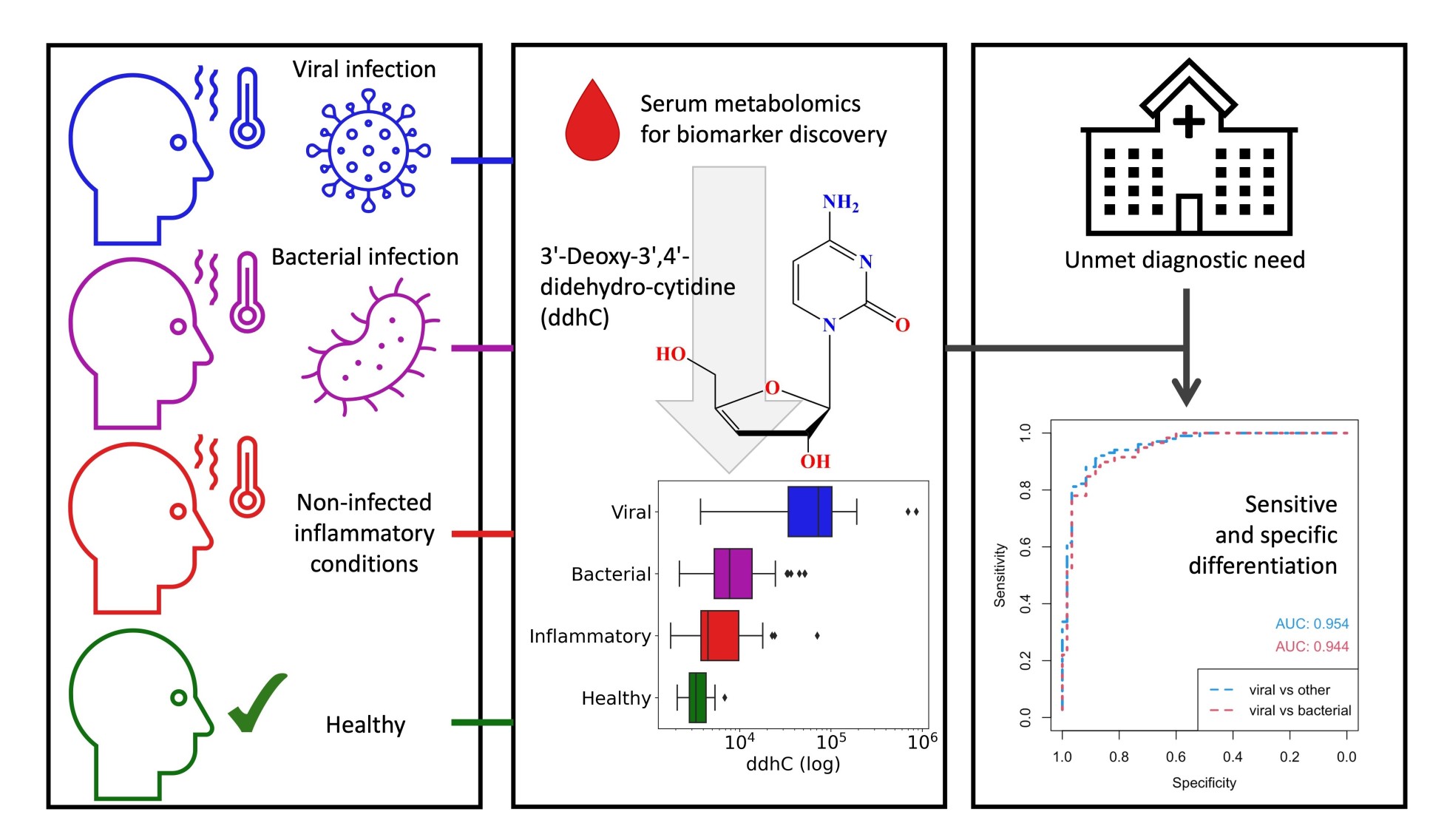
The study, led by academic clinical fellow Dr Ravi Mehta, found that serum from patients who had been admitted with suspected infection contained a novel small molecule known as ddhC, which is the by-product of the only known antiviral molecule that humans make.
COVID-19 has also shown us that there are specific advantages to recognising viral infection - from offering specific therapeutics, to isolating the patient and protecting others. Prof. Shiranee Sriskandan
Levels of ddhC were high in people who had an acute viral infection (whether COVID-19 or flu) but were not high in the other patients, including those with bacterial infections and those who turned out not to have an infection. These differences in levels are considered sufficient to use ddhC as a diagnostic test to support an acute viral infection.
The study, published in Med, is a collaboration between Imperial's Department of Infectious Disease and the National Phenome Centre (NPC), based in the Department of Metabolism, Digestion and Reproduction, as well as University College Hospital London, and was funded by the NIHR Imperial BRC.
Speaking about the importance of having a test for viral infection, senior author Professor Shiranee Sriskandan said: "Having a generic diagnostic test that distinguishes ‘viral infection from bacterial infection’ stemmed originally from a desire to help identify those patients presenting to the emergency department who would, or would not, benefit from an antibiotic, hence supporting the fight against antimicrobial resistance by avoiding over-prescribing.
"Everyone accepts that it's necessary to identify a bacterial diagnosis to ensure that we give the right antibiotic if required, but the same has not been true for viral infection. However, we increasingly recognise that making a diagnosis of a viral infection can be important; for example in the early stages of a pandemic or newly emerging febrile syndrome, a generic biomarker for viral infection might be very useful."
Metabolic profiling
Metabolic profiling technology was the key tool used in the discovery of this biomarker. The technique simultaneously measures hundreds and thousands of small molecule metabolites in human biofluids, such as blood products (serum and plasma) and urine, providing a “snapshot” of health obtained through the analysis of amino acids, lipids, sugars, and other metabolites under a particular condition.
Due to the broad chemical diversity and different concentration levels of metabolites present in human biofluids and tissues, metabolic profiling experiments require the use of a wide range of analytical methods and platforms to detect, isolate, identify and quantify various classes of metabolites.
For this project, the NPC employed a portion of its standard serum profiling platform utilising liquid chromatography-mass spectrometry (LC-MS) technology. A suite of assays was used to maximise the coverage of the metabolome.
The metabolic phenotyping technologies used here are powerful in their ability to discover new metabolites that are associated (in this case) with a particular disease phenotype from the thousands of metabolites measured. However, this creates a bottleneck, when signals are identified as biomarkers but their chemical identity remains unknown.
Speaking about the profiling process, Dr. Matt Lewis, Director of the NPC, said: "Our experienced team is dedicated to biomarker identification, ensuring the signals produced by the platform are immediately interpretable. In this case, the team employed tandem mass spectrometry to determine the chemical “fingerprint” of the biomarker and then deduced its likely composition and structure from that information. The annotation was confirmed by comparison of data produced using the same LC-MS techniques to that of a reference material, ensuring a confident identification."
The data analysis conducted by the teams, and the biomarkers arising, were identified as the novel metabolite 3'-Deoxy-3',4'-didehydro-cytidine (ddhC).
Study significance
On the importance of the study results, Dr Matt Lewis said: "From an “omics” perspective, the study has been a great success, especially because of its direct influence on improving clinical care. Rarely do single novel biomarkers emerge from these kinds of studies with such strong diagnostic potential. More often we are looking at more subtle effects across wider panels of metabolites. However here, there really appears to be a single “silver-bullet” biomarker that indicates viral infection, which makes sense given its chemical relation to the only known human antiviral small molecule. The results are really significant because of the biomarker’s immediate potential for use in rapid diagnostics for viral infection."
This work has been successfully awarded Confidence In Concept funding to progress the result towards a clinically translational endpoint of a potential diagnostic test.
Funded by the NIHR Imperial BRC, the study was undertaken using patient samples from Imperial’s Infection Bioresource and the BioAID study, which are housed in the BRC’s Leonard and Dora Colebrook laboratory.
Antiviral metabolite 3′-deoxy-3′,4′-didehydro-cytidine is detectable in serum and identifies acute viral infections including COVID-19. Ravi Mehta, Elena Chekmeneva, Heather Jackson, Caroline Sands, Ewurabena Mills, Dominique Arancon, Ho Kwong Li, Paul Arkell, Timothy M. Rawson, Robert Hammond, Maisarah Amran, Anna Haber, Graham S. Cooke, Mahdad Noursadeghi, Myrsini Kaforou, Matthew R. Lewis, Zoltan Takats, Shiranee Sriskandan. MED. January 27, 2022. DOI: https://doi.org/10.1016/j.medj.2022.01.009
Article supporters
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Benjie Coleman
Department of Surgery & Cancer