Decoding cellular conversations in ulcerative colitis reveals new drug targets
New research reveals previously hidden mechanisms driving inflammation and identifies new therapeutic targets, including some already showing success in late-stage clinical trials.
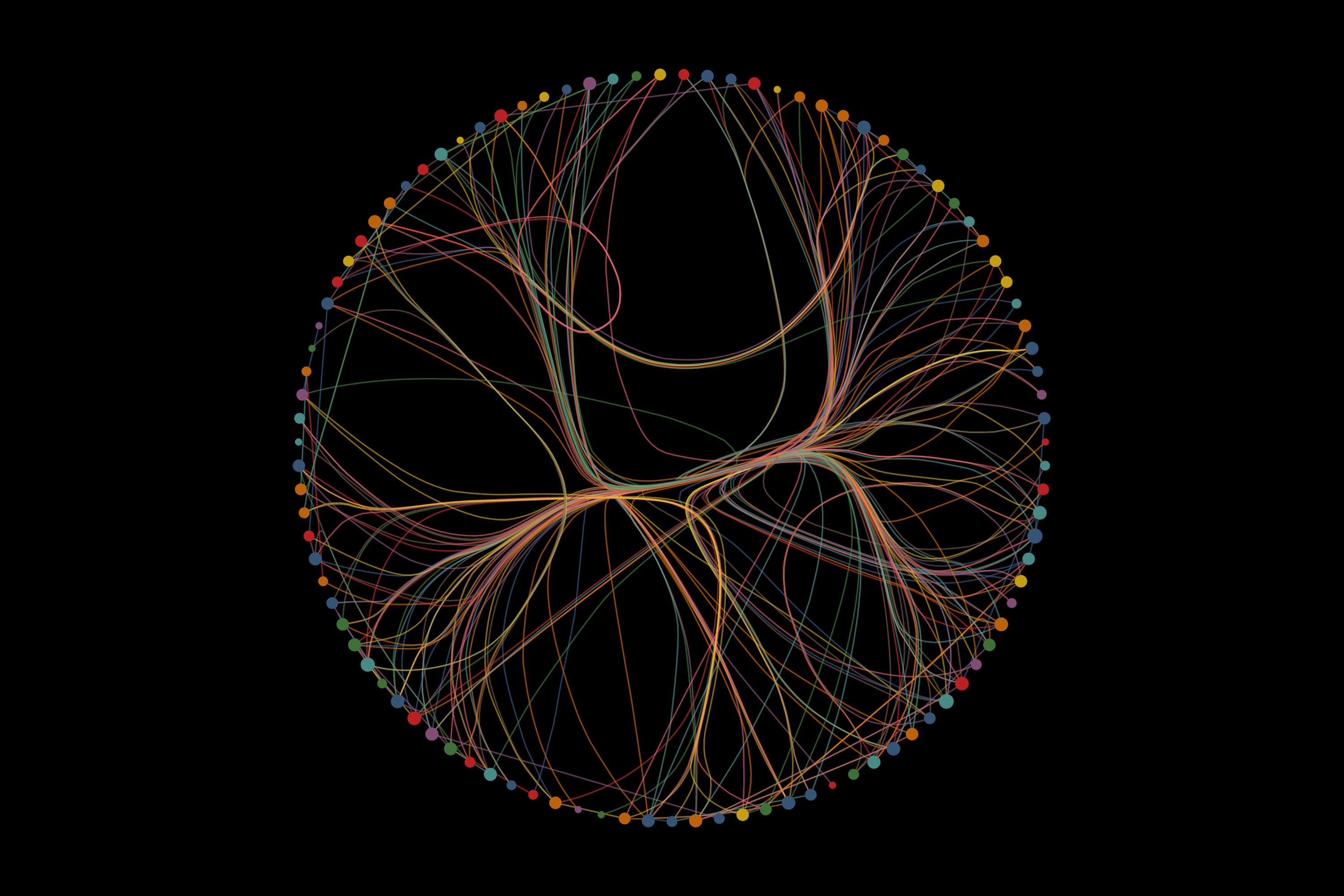
We moved beyond studying individual cytokines, and mapped them as an interacting network to reveal, for the first time, how this immune regulation is organised in healthy tissue, disrupted in ulcerative colitis, and altered by therapy. Dr Marton Olbei Imperial Teaching Fellow and study first author
Imperial researchers have, for the first time, mapped how inflammatory cytokines regulate one another to form coordinated signalling networks in healthy tissue and ulcerative colitis, revealing how this immune regulatory blueprint is rewired in disease.
The study, led by Associate Professor Tamas Korcsmaros from the Department of Metabolism, Digestion and Reproduction, not only reveals previously hidden mechanisms driving inflammation, but also identifies new therapeutic targets, including some already showing success in late-stage clinical trials.
By revealing how inflammatory signals are organised across different cell types, the research provides a framework for identifying more effective drug targets and for understanding why some treatments succeed while others fail.
Published in Science Signaling, Decoding cytokine networks in ulcerative colitis to identify pathogenic mechanisms and therapeutic targets, represents a major advance in understanding inflammatory bowel disease at a systems level.
Listening in on cellular communication
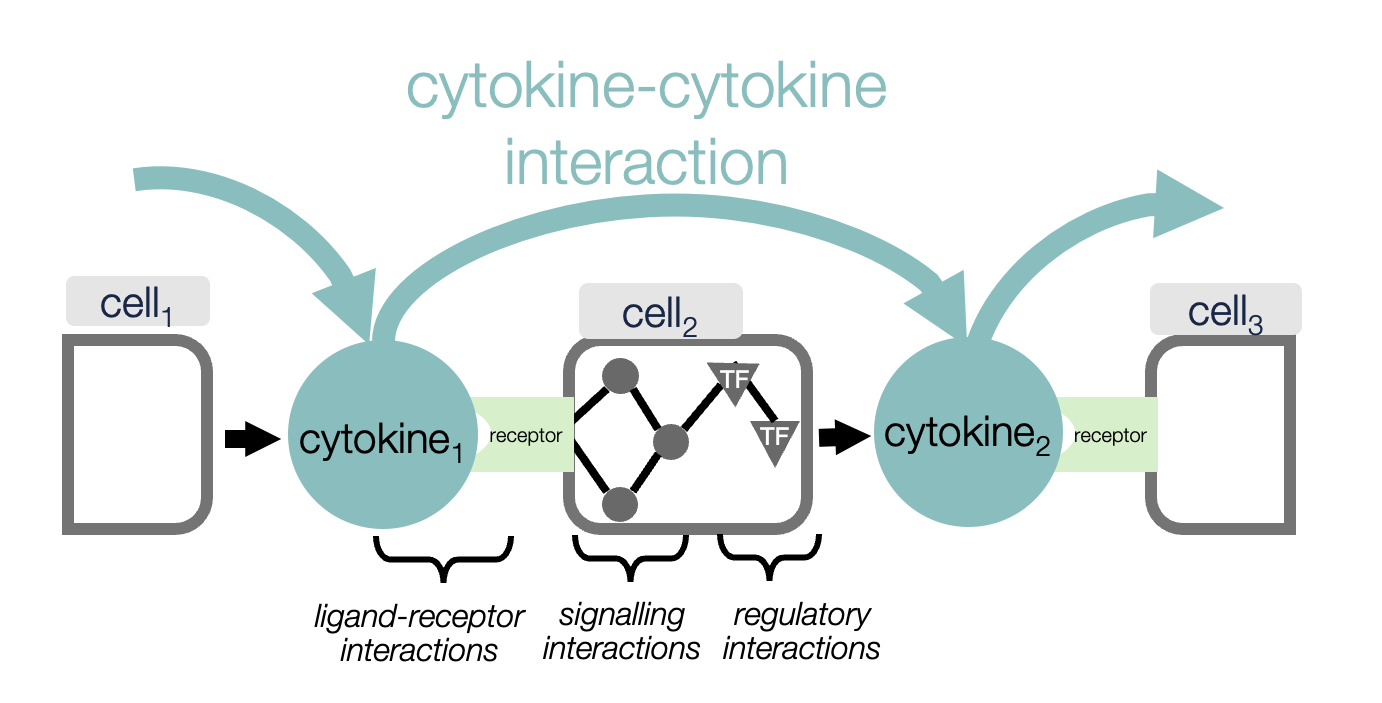
Ulcerative colitis (UC), a major form of inflammatory bowel disease, is caused by abnormal immune activity in the gut. Many current treatments target individual inflammatory cytokines, but up to 70% of patients either do not respond or stop responding over time. One reason is that these molecules do not work alone, they are part of complex communication networks linking immune, gut lining and support cells.
While individual cytokines and their effects have been studied extensively, it has not previously been possible to map how cytokines regulate each other as a coordinated network, or to see how this network differs between health, disease and treatment.
This research combined single-cell RNA sequencing data from colonic biopsies of more than 120 individuals with advanced computational modelling to reconstruct cytokine-cytokine interaction networks. These networks reveal how one cytokine triggers the production of others across different cell types, forming cascades of inflammatory signalling that define the immune state of healthy tissue or disease.
Speaking about the findings, Dr Marton Olbei, Imperial Teaching Fellow and first author of the study, said: “We moved beyond studying individual cytokines, and mapped them as an interacting network to reveal, for the first time, how this immune regulation is organised in healthy tissue, disrupted in ulcerative colitis, and altered by therapy.”
Organoid validation and collaboration
The researchers validated their findings using patient-derived intestinal organoids, three-dimensional mini-guts grown from UC patients, as well as organoid-immune cell co-culture systems.
Dr Tamas Korcsmaros, Imperial Organoid Facility lead and co-senior author of the study, said: “We didn’t just predict new biology using computational models but validated these predictions experimentally using organoids derived directly from patients.”
This combination is scientifically powerful and highlights the growing role the NIHR Imperial BRC Organoid Facility plays in demonstrating how organoids can bridge the gap between big data analysis and clinically relevant biology.
This project has been the first to benefit from the Imperial co-founded LION - the London gastroIntestinal Organoid Network, which got established recently. The organoid-immune co-culture system experiment was made possible by colleagues at King’s College London to complement Imperial's efforts and locally available expertise.
A blueprint for future therapies
Importantly, as this study not just established the approach but also validated it for one type of inflammatory diseases, there is an exciting opportunity to apply the same methodology for other complex diseases where cytokines play a key role.
Dr Dezső Módos, Imperial College Research Fellow and co-senior author of the study, said: "Finding the right treatment for the right patient is a major challenge for clinicians. With this new framework we can understand better how the communication between key cells is changing, and this will enable us to predict in a computer how certain treatments work and why some patients don't respond to therapies."
This new study, and its published methods, will open the way for the community to carry out similar analyses in other diseases, spanning from inflammatory conditions such as rheumatoid arthritis, to infections and to currently hard to treat diseases like chronic fatigue syndrome.
Marton Olbei et al, Decoding cytokine networks in ulcerative colitis to identify pathogenic mechanisms and therapeutic targets. Sci. Signal. 19, eadt0986(2026). DOI: 10.1126/scisignal.adt0986
Infrastructure support for this research was provided by the NIHR Imperial Biomedical Research Centre (BRC).
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Article people, mentions and related links
Benjie Coleman
Faculty of Medicine
