Characterising disease progression through feature landscapes and clusters of symptoms
Prof Mauricio Barahona (Mathematics) and Prof Paul Matthews (Medicine)
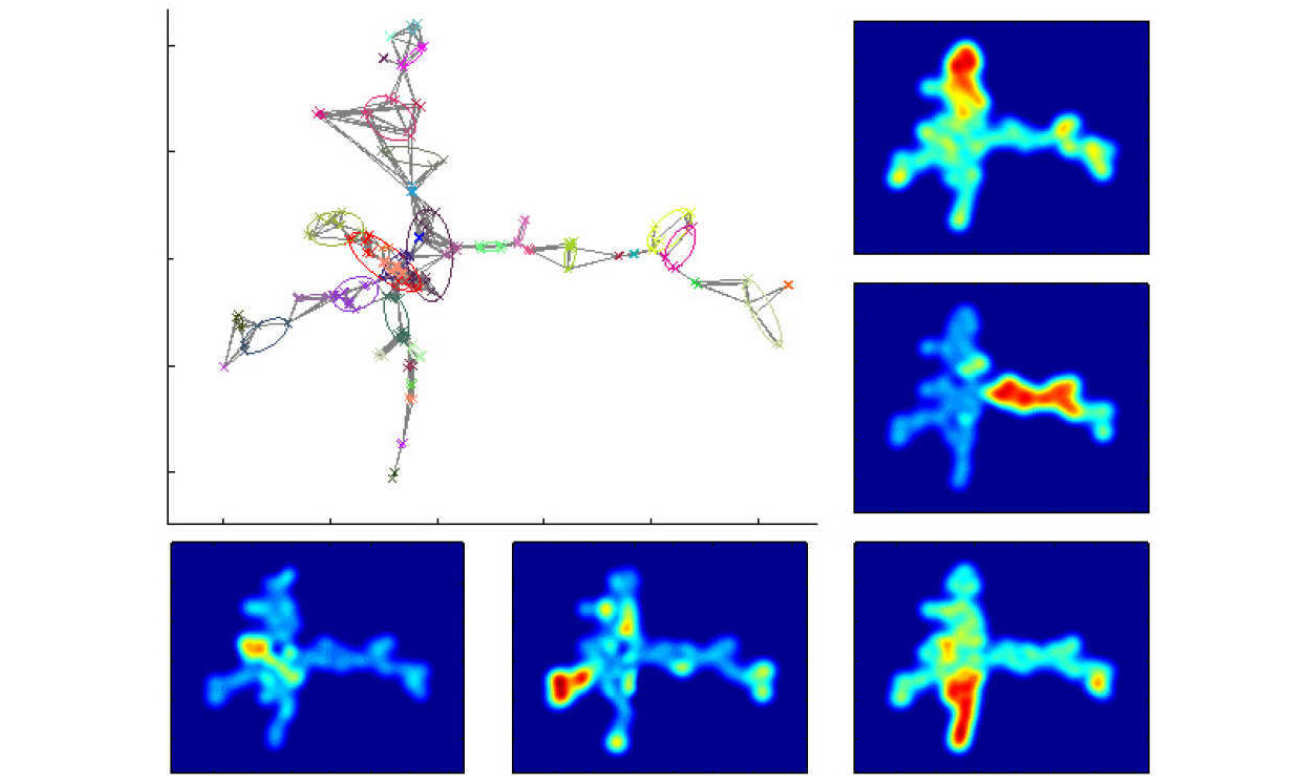
This project is aimed at developing mathematical tools for the analysis of high-dimensional datasets characterising disease nosology and progression. The tools involve graph-theoretical approaches to dimensionality reduction and coarse-graining of spatio-temporal landscapes using stochastic processes. The objective is to obtain feature landscapes describing multiple factors of disease progression and to cluster symptoms and patients using community detection algorithms. Our hypothesis is that clusters or assemblies of symptoms can serve us to explore similarities (and potentially shared mechanisms) between disorders that were previously viewed as unrelated. The project will use large datasets from the UK Biobank and Dementia Platforms UK, which include both case control studies with well phenotyped subjects as well as high-dimensional quantitative biological characterisations (including epidemiological cohort data and genetics).
Image shows the patterns of gene expression in the process of differentiation of hematopoietic stem cells occurs on a high-dimensional transcriptomic landscape, captured here through geometric graphs and dimensionality reduction. The features of the landscape can be clustered to characterise the different branches of cell type progression.
The Team
The team
Prof Mauricio Barahona

Prof Mauricio Barahona
Mathematics
Prof Paul Matthews

Prof Paul Matthews
Medicine
--tojpeg_1538046453332_x4.jpg)
