CRISPR-based oligo recombineering in drug discovery
Congratulations to Henry Benns for his first-author publication in Nature Microbiology
Small-molecule drugs that inhibit the function of proteins in cells can be powerful in stemming pathogenic infections and disease. However, creating these drugs relies on finding the best sites in proteins for them to bind to.
Amino acids are the building blocks of proteins, and their sequence defines protein function. Therefore, finding specific amino acids that bind with potential drug molecules and impact function is vital. However, protein sequence function relationships are difficult to predict.
This research, led by Dr Matt Child and in close collaboration with Prof. Ed Tate, has created a way to systematically quantify the contribution of hundreds of druggable amino acids to protein function in live cells. Details of this technology can be found in our Nature Microbiology article.
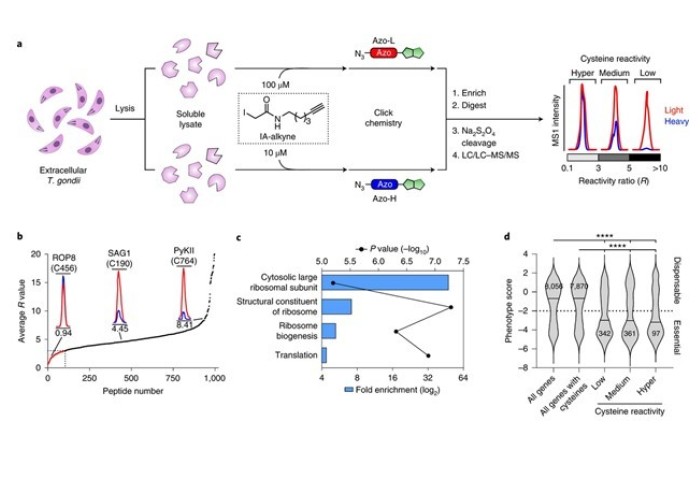
They tested their system, called CRISPR-based Oligo Recombineering (CORe), in the parasite Toxoplasma gondii. They discovered specific cysteine amino acids decorating the ribosome that were critical for protein function and parasite growth.
Using this information, they undertook a screen of drug-like molecules to hunt for cysteine-targeted drugs that inhibited ribosome function. This identified a first-in-class parasite-selective antimalarial molecule that inhibited protein translation, which could be repurposed to target T. gondii.
Well done to everyone involved! A huge thank you to all our collaborators and funding bodies for their support.
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Dr Ravi Singh
Department of Chemistry