AI for life sciences research gets boost from the financial markets
A technique to find hidden connections between stocks and shares has been applied to discovering and mapping connections between proteins.
The method aims to speed up life sciences research by automatically updating knowledge about and finding new correlations between genes, proteins and small molecules. Many of these are involved in disease and could be targets for new drugs.
Proteins play a role in all the essential functions of our cells, so mapping their interactions, and which other molecules modify these interactions, is of huge importance. Dr Sarah Rouse
The technique uses a branch of artificial intelligence (AI) called ‘natural language understanding’ to scan documents for connections between concepts. In the financial sector, it can be used to find correlations between certain stocks and shares and how they change over time, allowing companies to invest or divest before others are aware of a change.
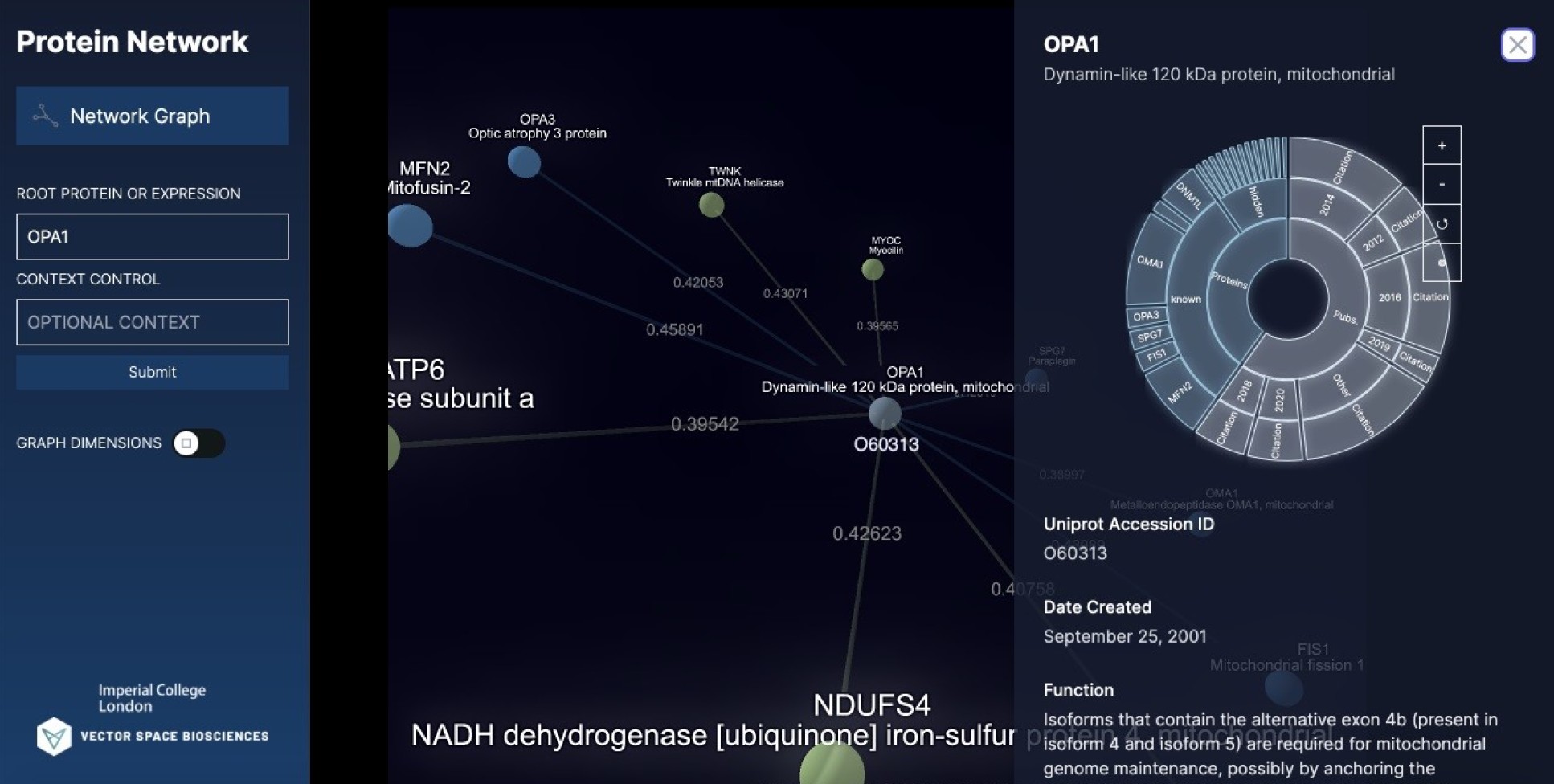
Now, Imperial College London and Vector Space Biosciences researchers have applied the technique to the academic literature, providing a new way to quickly map the changing information about how proteins relate to each other.
Scanning the literature manually for updates can take a lot of a researcher’s time, and large databases can soon be out of date. The new technique updates information about proteins’ relationships in real time, ensuring researchers are working with the most up-to-date information.
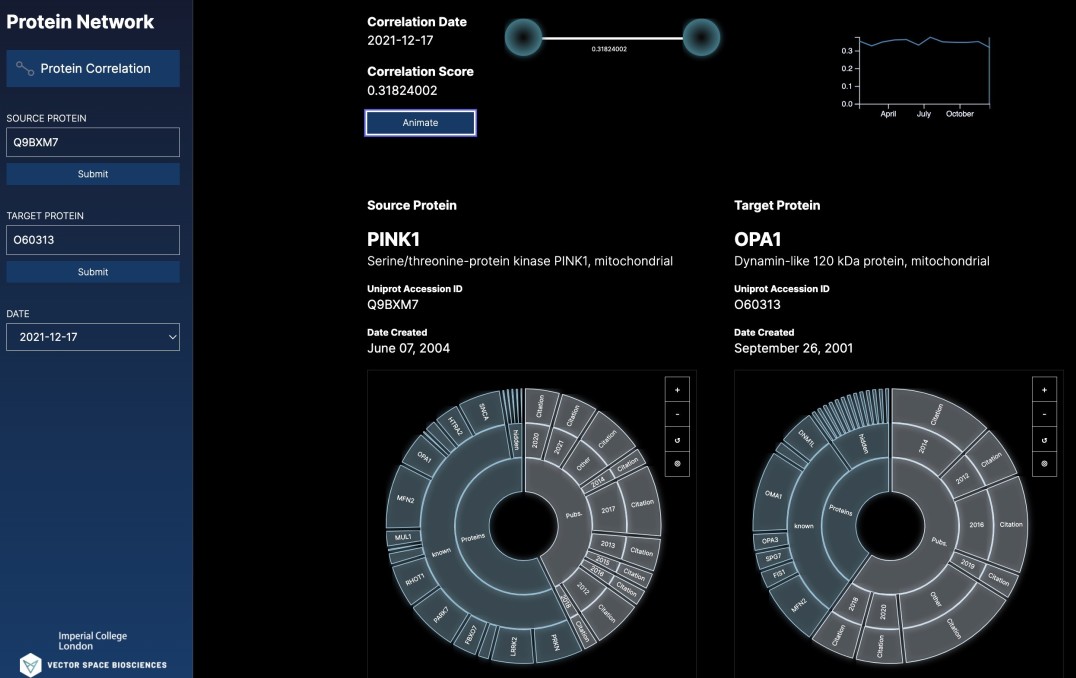
The first results of the collaboration, a map showing the evolution of the observed correlation between two proteins over time, is being unveiled at The Biophysical Society annual meeting in San Francisco.
Ageing proteins
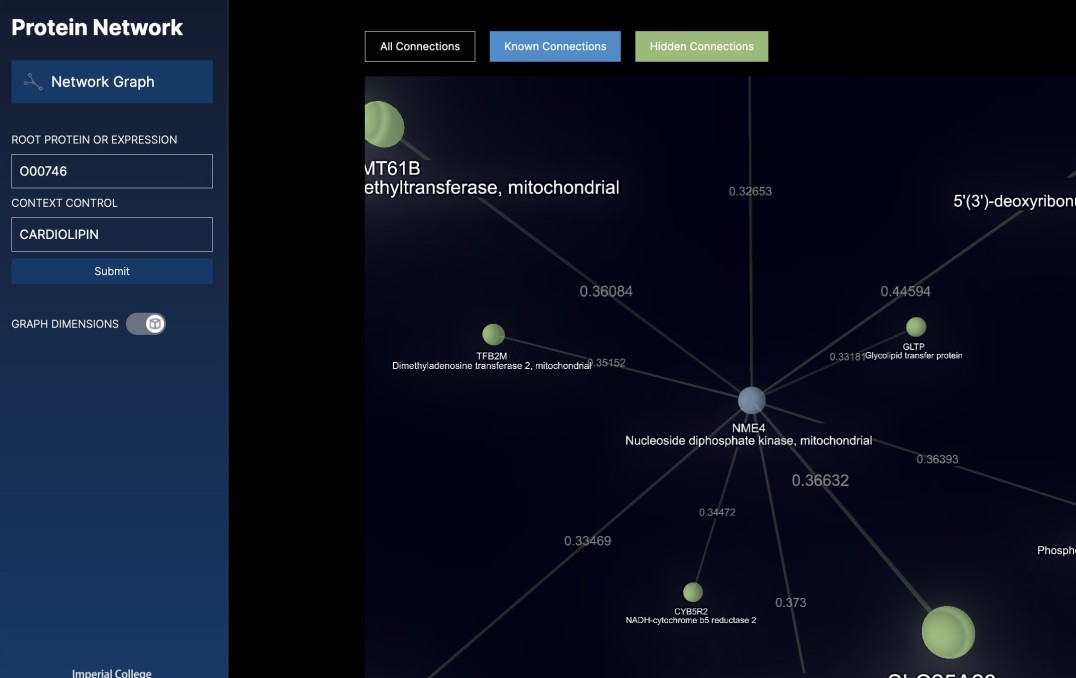
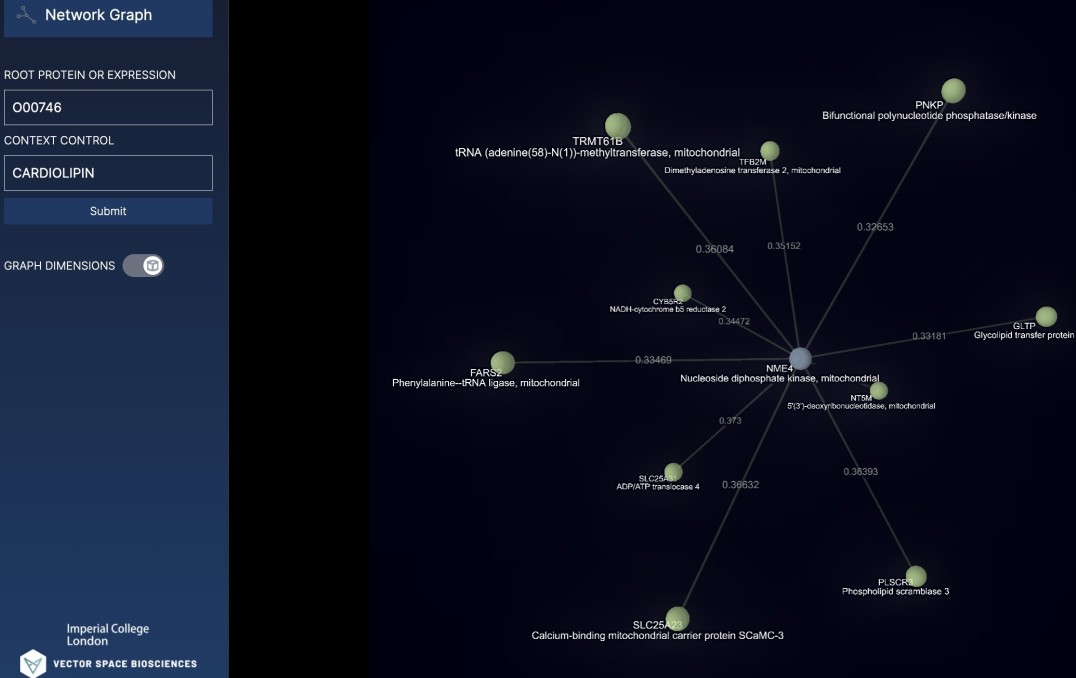
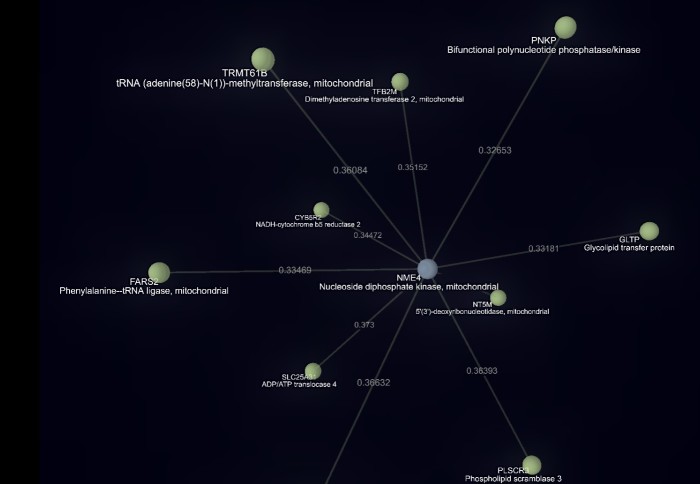
Dr Sarah Rouse, from the Department of Life Sciences at Imperial, is particularly interested in how proteins interact via a lipid molecule called cardiolipin. Cardiolipin mediates the interaction between proteins in the membranes of mitochondria – the powerhouses of cells.
Cardiolipin is known to be important in ageing, as well as in age-related disorders including neurodegenerative disease, cardiomyopathies (heart-related diseases) and cancer. But only by knowing the detailed mechanisms of how cardiolipin affects proteins and their related actions in ageing can researchers hope to design potential new drug molecules to slow any degenerative processes.
For this, researchers need the most up-to-date information about protein interactions. The new AI technique captures this but also goes a step further – by looking for ‘hidden’ relationships in the literature. These may be ways the proteins interact that are not explicitly stated in any research papers, but that the AI can infer by analysing the language.
Dr Rouse said: “Fundamental biology studies help us understand what the body does at a molecular level when it’s healthy, which can lead us to understand what goes wrong in disease. Proteins play a role in all the essential functions of our cells, so mapping their interactions, and which other molecules modify these interactions, is of huge importance.
“However, it’s also a huge challenge. While there is much research and data being produced, how it all fits together can sometimes be difficult to keep up with. Now, with the techniques developed by Vector Space Biosciences we have been able to streamline this process and discover ‘hidden connections’ – those that, for example, would not be made without a researcher trawling the literature and making the connection manually.”
For Dr Rouse and her team at Imperial, this new efficiency will be crucial in accelerating their research on how lipids in particular modify protein connections. Lipids are not currently popular drug targets, but investigating their action in this way could open up the pathway to a whole new class of lipid-based drugs.
Link to spaceflight
Vector Space Biosciences (VSB) is particularly interested in the research as it relates to ageing, and relatedly how the body responds to spaceflight, which has been shown in an astronaut twin study to affect the same ageing processes in cells.
VSB got their start in space biosciences in the early 2000s, using language modelling to detect hidden relationship networks between genes, proteins and drug compounds in the context of human ageing, chromosomal radiation damage and breast cancer.
They then used these same methods to detect hidden relationships between stocks, cryptos and global events in the financial markets. CEO of VSB Kasian Franks said: “We're excited to be collaborating with the Rouse Lab at Imperial as their research connects to the core of what we do in terms of developing countermeasures for stressors to protect and repair the human body during spaceflight with benefits to all humankind."
Undergraduate Chemistry student Kin Chao, who has been contributing to the project, said: "I have really enjoyed the opportunity to work with industry partners VSB as part of a multidisciplinary research experience.”
Article text (excluding photos or graphics) © Imperial College London.
Photos and graphics subject to third party copyright used with permission or © Imperial College London.
Reporter
Hayley Dunning
Communications Division