Integrating imaging and next generation sequencing to characterise disease states using geometric and topological graph-based dimensionality reduction
Prof Daniel Rueckert (Computing), Prof Mauricio Barahona (Mathematics)
Others: Prof Paul Matthews (Medicine), Dr Declan O’Regan, Prof Stuart Cook (MRC Clinical Sciences Centre and BHF Centre, Medicine), Dr Colin Cotter (Mathematics), Dr Mariano Beguerisse (Mathematical Institute, University of Oxford), Dr Heather Harrington (Mathematical Institute, University of Oxford)
With the advent of advanced 3D imaging and next generation sequencing, there is a pressing need to integrate molecular, genetic and spatio-temporal imaging data to guide diagnostics and the initiation of therapeutic agents. The Imperial Biomedical Imaging group has ample expertise in advanced medical imaging with well-developed protocols for clinical settings. Data has been collected on medical cohorts both for cardiomyopathies as well as neurodegenerative diseases.
The mathematics of manifold learning provides a point of contact between advanced image processing techniques and feature-based analysis of molecular and genetic data. In both cases, dimensionality reduction can be formulated in terms of sparsification and coarse-graining of geometric graphs derived from images and molecular data, leading to similarity graphs based on feature vectors that combine spatial, morphological and functional data from high dimensional imaging (e.g., high resolution 3D MRI) with genetic and molecular data associated with disease. Sparsification and coarse-graining of those graphs will be sought to obtain reduced sets of relevant features for the description of controlled cohorts. The methods will build upon graph-theoretical partitioning tools based on generalized Laplacians and simplicial complexes, graph-partitioning for geometric dimensionality reduction, and techniques from topological data analysis to enhance robustness. Currently used statistical models of anatomy and function across populations of subjects will thus be enriched with phenotypic feature information. Advanced methodologies using shape descriptors and localized image markers will also be explored. Medical scientists will be able to review the results using intuitive online decision-support tools targeted at the clinic to guide individualised management based on current platforms in use at Imperial.
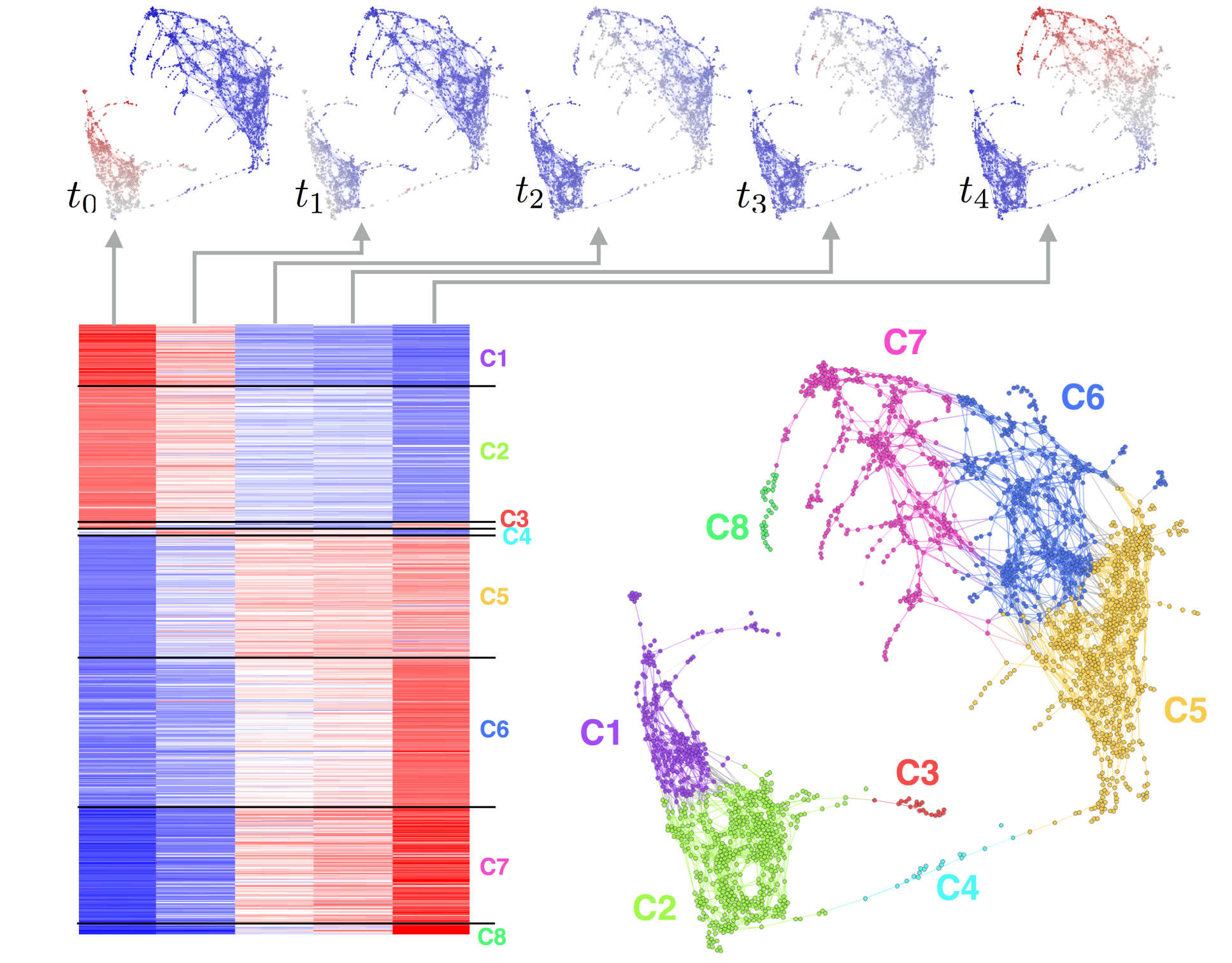
Clustering gene transcription temporal patterns observed during developmental stages using geometric graphs and Markov processes
The Team
Leaders
Prof Daniel Rueckert
/prod01/channel_3/media/migration/research-groups/Rueckert--tojpeg_1469627098462_x4.jpg)
Prof Daniel Rueckert
Computing
Prof Mauricio Barahona
/prod01/channel_3/media/migration/research-groups/barahona--tojpeg_1469627218200_x4.jpg)
Prof Mauricio Barahona
Mathematics
Others
Prof Paul Matthews
/prod01/channel_3/media/migration/research-groups/matthews--tojpeg_1469627377259_x4.jpg)
Prof Paul Matthews
Medicine
Dr Declan O’Regan
/prod01/channel_3/media/migration/research-groups/oregan--tojpeg_1469627466343_x4.jpg)
Dr Declan O’Regan
Medicine
Prof Stuart Cook
/prod01/channel_3/media/migration/research-groups/cook--tojpeg_1469627537525_x4.jpg)
Prof Stuart Cook
Medicine
Dr Colin Cotter
/prod01/channel_3/media/migration/research-groups/cotter--tojpeg_1469627608054_x4.jpg)
Dr Colin Cotter
Mathematics
Dr Mariano Beguerisse
/prod01/channel_3/media/migration/research-groups/picMBD--tojpeg_1469714026235_x4.jpg)
Dr Mariano Beguerisse
Mathematical Institute, Oxford
Dr Heather Harrington
/prod01/channel_3/media/migration/research-groups/harrington--tojpeg_1469714055443_x4.jpg)
Dr Heather Harrington
Mathematical Institute, Oxford
--tojpeg_1538046453332_x4.jpg)
