We work on a number of enteric pathogens, including poliovirus, rotavirus, enteroviruses, typhoid, Clostridium difficile and Campylobacter. We are also using next-generation sequencing methods to characterise the intestinal microbiome (viruses, bacteria, fungi) and its relationship with host immunity.
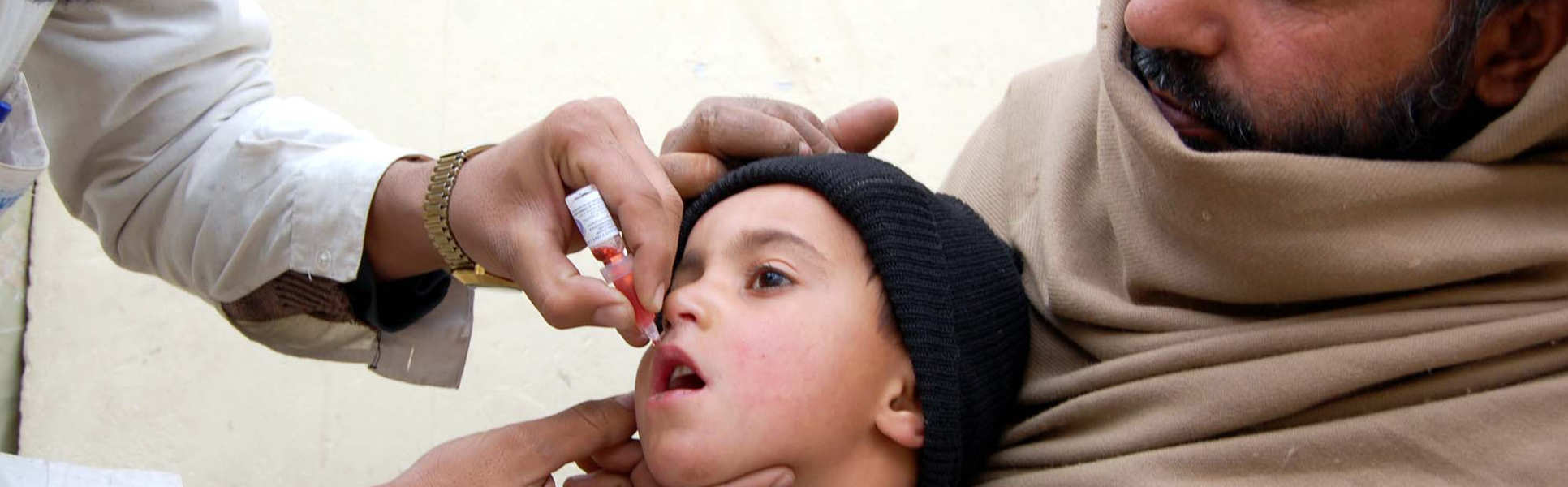
Our research on poliovirus has led to new uses of polio vaccines and informed global polio eradication strategy and planning. With partners in Asia and Africa, we have conducted epidemiological studies and clinical trials to estimate the immunogenicity and effectiveness of different oral poliovirus vaccines and of inactivated poliovirus vaccine given as a booster dose. We are currently working on vaccination strategies to mitigate risks associated with global withdrawal of oral poliovirus vaccine and on next-generation sequencing of sewage samples to detect and characterise circulating polioviruses.
We are working with collaborators in the UK, India and Africa to identify the causes of poor oral rotavirus vaccine immunogenicity and efficacy in low-income settings. This work is focused on the intestinal microbiome and maternal factors including breastmilk antibodies and histo-blood group antigens.
With collaborators in the UK and Europe, we are using genetic sequencing technologies to elucidate the origins and spread of pathogenic enteric bacteria including Salmonella, Clostridium difficile and Campylobacter jejuni. Current research is focused on methods to analyse next-generation sequencing data and to model the spread of antimicrobial resistance.
We are also working on typhoid epidemiology and surveillance with collaborators in India and Vietnam and on typhoid serology assays.
People
Professor Nicholas Grassly
/prod01/channel_3/media/migration/under-construction/Nicholas_Grassly--tojpeg_1479727261602_x4.jpg)
Professor Nicholas Grassly
Professor of Infectious Disease & Vaccine Epidemiology
Dr Isobel Blake
/prod01/channel_3/media/migration/faculty-of-medicine/Isobel-Blake--tojpeg_1543592601293_x4.jpg)
Dr Isobel Blake
Lecturer
Professor Christl Donnelly
/prod01/channel_3/media/migration/faculty-of-medicine/Christl_Donnelly--tojpeg_1479727046231_x4.jpg)
Professor Christl Donnelly
Professor of Statistical Epidemiology
Dr Thibaut Jombart
/prod01/channel_3/media/migration/faculty-of-medicine/Thibaut_Jombert--tojpeg_1479730493731_x4.jpg)
Dr Thibaut Jombart
Senior Lecturer
Contact us
For any enquiries related to the MRC Centre please contact:
Scientific Manager
Susannah Fisher
mrc.gida@imperial.ac.uk
External Relationships and Communications Manager
Dr Sabine van Elsland
s.van-elsland@imperial.ac.uk