Single cell genomics
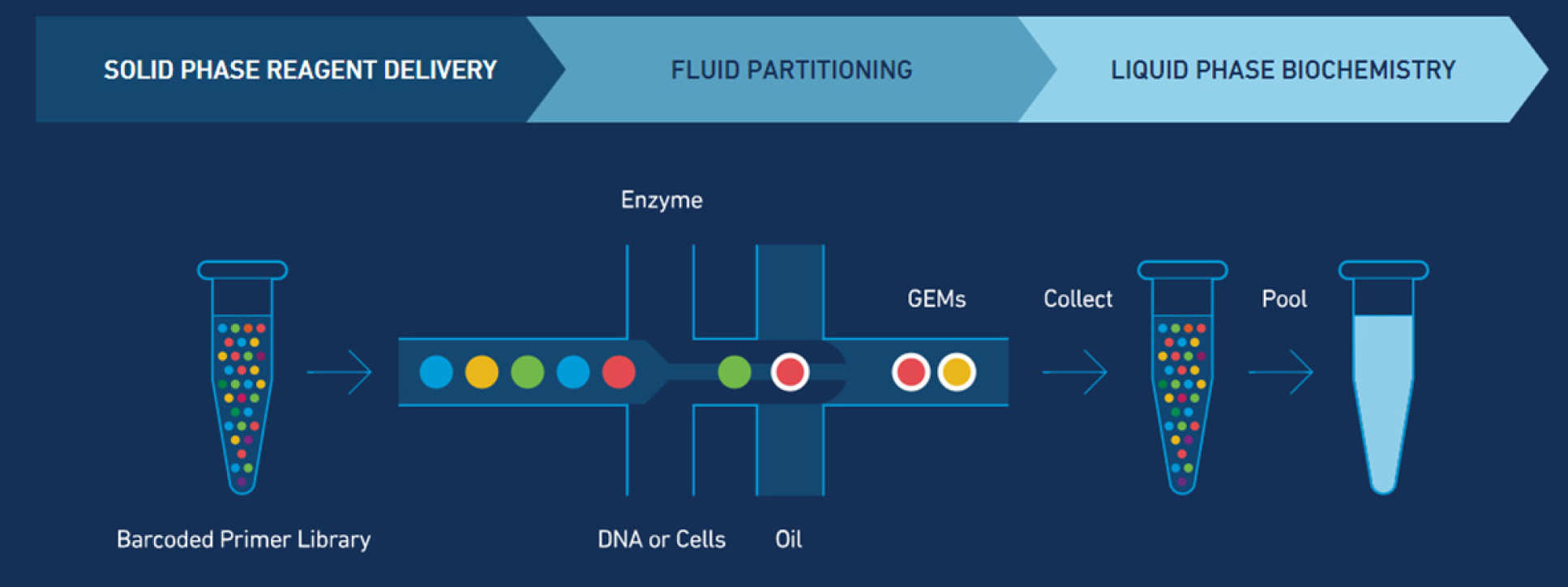
10X Genomics Single Cell Chromium, modified from https://www.10xgenomics.com/technology/
The Chromium Single Cell 3′ platform is now available within the BRC Genomics Facility. The system provides a comprehensive, scalable solution for single cell genomics profiling of thousands of cells with each preparation. Up to eight independent samples can be quickly processed in parallel. The instrument uses microfluidics technology to generate an emulsion in which droplets encapsulate a cell and reagents. Barcoded oligos are used in each droplet to assign a cell-specific tag to each captured mRNA molecule. The cDNA produced is used to generate a 3'end RNA library for Illumina sequencing. After sequencing, the cell-specific barcodes are used to separate the reads on a per-cell fashion, additional unique molecular identifiers (UMI) are used to remove PCR duplicates.
Loupe browser and report

10X Genomics' Loupe Cell Browser
10X Genomics' Loupe Cell Browser can be freely downloaded to visualise data files as well as:
- Identify genes that uniquely characterize clusters
- Locate different cell types and functional groups based on gene expression
- Create new clusters and dissect your data in even smaller subgroups
- Save genes of interest, export data tables, and capture screenshots of your single-cell data
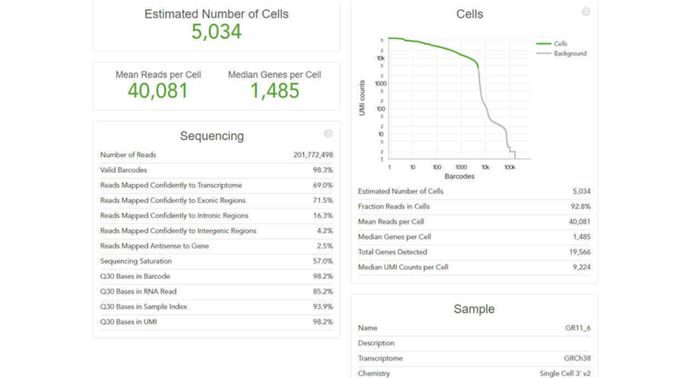
Example of report
- Example of a report showing the number of cells detected, number of reads and genes per cell.
- Additional metrics are shown including sequence saturation and data quality.


